A novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI), GRL-98065, is potent against multiple-PI-resistant human immunodeficiency virus in vitro
- PMID: 17371811
- PMCID: PMC1891396
- DOI: 10.1128/AAC.01413-06
A novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI), GRL-98065, is potent against multiple-PI-resistant human immunodeficiency virus in vitro
Abstract
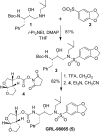
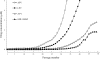
We designed, synthesized, and identified GRL-98065, a novel nonpeptidic human immunodeficiency virus type 1 (HIV-1) protease inhibitor (PI) containing the structure-based designed privileged cyclic ether-derived nonpeptide P2 ligand, 3(R),3a(S),6a(R)-bis-tetrahydrofuranylurethane (bis-THF), and a sulfonamide isostere, which is highly potent against laboratory HIV-1 strains and primary clinical isolates (50% effective concentration [EC(50)], 0.0002 to 0.0005 microM) with minimal cytotoxicity (50% cytotoxicity, 35.7 microM in CD4(+) MT-2 cells). GRL-98065 blocked the infectivity and replication of each of the HIV-1(NL4-3) variants exposed to and selected by up to a 5 microM concentration of saquinavir, indinavir, nelfinavir, or ritonavir and a 1 microM concentration of lopinavir or atazanavir (EC(50), 0.0015 to 0.0075 microM), although it was less active against HIV-1(NL4-3) selected by amprenavir (EC(50), 0.032 microM). GRL-98065 was also potent against multiple-PI-resistant clinical HIV-1 variants isolated from patients who had no response to existing antiviral regimens after having received a variety of antiviral agents, HIV-1 isolates of various subtypes, and HIV-2 isolates examined. Structural analyses revealed that the close contact of GRL-98065 with the main chain of the protease active-site amino acids (Asp29 and Asp30) is important for its potency and wide-spectrum activity against multiple-PI-resistant HIV-1 variants. The present data demonstrate that the privileged nonpeptide P2 ligand, bis-THF, is critical for the binding of GRL-98065 to the HIV protease substrate binding site and that this scaffold can confer highly potent antiviral activity against a wide spectrum of HIV isolates.
Figures
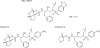


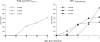

Similar articles
-
GRL-02031, a novel nonpeptidic protease inhibitor (PI) containing a stereochemically defined fused cyclopentanyltetrahydrofuran potent against multi-PI-resistant human immunodeficiency virus type 1 in vitro.Antimicrob Agents Chemother. 2009 Mar;53(3):997-1006. doi: 10.1128/AAC.00689-08. Epub 2008 Oct 27. Antimicrob Agents Chemother. 2009. PMID: 18955518 Free PMC article.
-
GRL-0519, a novel oxatricyclic ligand-containing nonpeptidic HIV-1 protease inhibitor (PI), potently suppresses replication of a wide spectrum of multi-PI-resistant HIV-1 variants in vitro.Antimicrob Agents Chemother. 2013 May;57(5):2036-46. doi: 10.1128/AAC.02189-12. Epub 2013 Feb 12. Antimicrob Agents Chemother. 2013. PMID: 23403426 Free PMC article.
-
Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro.Antimicrob Agents Chemother. 2003 Oct;47(10):3123-9. doi: 10.1128/AAC.47.10.3123-3129.2003. Antimicrob Agents Chemother. 2003. PMID: 14506019 Free PMC article.
-
Design of HIV protease inhibitors targeting protein backbone: an effective strategy for combating drug resistance.Acc Chem Res. 2008 Jan;41(1):78-86. doi: 10.1021/ar7001232. Epub 2007 Aug 28. Acc Chem Res. 2008. PMID: 17722874 Review.
-
Darunavir: a review of its use in the management of HIV infection in adults.Drugs. 2009;69(4):477-503. doi: 10.2165/00003495-200969040-00007. Drugs. 2009. PMID: 19323590 Review.
Cited by
-
A Small Molecule, ACAi-028, with Anti-HIV-1 Activity Targets a Novel Hydrophobic Pocket on HIV-1 Capsid.Antimicrob Agents Chemother. 2021 Sep 17;65(10):e0103921. doi: 10.1128/AAC.01039-21. Epub 2021 Jul 6. Antimicrob Agents Chemother. 2021. PMID: 34228546 Free PMC article.
-
HIV-Associated Neurocognitive Disorder (HAND) and the Prospect of Brain-Penetrating Protease Inhibitors for Antiretroviral Treatment.Med Res Arch. 2017 Apr;5(4):1113. Epub 2017 Apr 15. Med Res Arch. 2017. PMID: 29984302 Free PMC article.
-
Non-cleavage site gag mutations in amprenavir-resistant human immunodeficiency virus type 1 (HIV-1) predispose HIV-1 to rapid acquisition of amprenavir resistance but delay development of resistance to other protease inhibitors.J Virol. 2009 Apr;83(7):3059-68. doi: 10.1128/JVI.02539-08. Epub 2009 Jan 28. J Virol. 2009. PMID: 19176623 Free PMC article.
-
Novel Protease Inhibitors Containing C-5-Modified bis-Tetrahydrofuranylurethane and Aminobenzothiazole as P2 and P2' Ligands That Exert Potent Antiviral Activity against Highly Multidrug-Resistant HIV-1 with a High Genetic Barrier against the Emergence of Drug Resistance.Antimicrob Agents Chemother. 2019 Jul 25;63(8):e00372-19. doi: 10.1128/AAC.00372-19. Print 2019 Aug. Antimicrob Agents Chemother. 2019. PMID: 31085520 Free PMC article.
-
A Modified P1 Moiety Enhances In Vitro Antiviral Activity against Various Multidrug-Resistant HIV-1 Variants and In Vitro Central Nervous System Penetration Properties of a Novel Nonpeptidic Protease Inhibitor, GRL-10413.Antimicrob Agents Chemother. 2016 Nov 21;60(12):7046-7059. doi: 10.1128/AAC.01428-16. Print 2016 Dec. Antimicrob Agents Chemother. 2016. PMID: 27620483 Free PMC article.
References
-
- Carr, A. 2003. Toxicity of antiretroviral therapy and implications for drug development. Nat. Rev. Drug Discov. 2:624-634. - PubMed
-
- De Clercq, E. 2002. Strategies in the design of antiviral drugs. Nat. Rev. Drug Discov. 1:13-25. - PubMed
-
- Erickson, J. W., and S. K. Burt. 1996. Structural mechanisms of HIV drug resistance. Annu. Rev. Pharmacol. Toxicol. 36:545-571. - PubMed
-
- Friesner, R. A., J. L. Banks, R. B. Murphy, T. A. Halgren, J. J. Klicic, D. T. Mainz, M. P. Repasky, E. H. Knoll, M. Shelley, J. K. Perry, D. E. Shaw, P. Francis, and P. S. Shenkin. 2004. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47:1739-1749. - PubMed
-
- Fumero, E., and D. Podzamczer. 2003. New patterns of HIV-1 resistance during HAART. Clin. Microbiol. Infect. 9:1077-1084. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

