The repressor element 1-silencing transcription factor regulates heart-specific gene expression using multiple chromatin-modifying complexes
- PMID: 17371849
- PMCID: PMC1900017
- DOI: 10.1128/MCB.00269-07
The repressor element 1-silencing transcription factor regulates heart-specific gene expression using multiple chromatin-modifying complexes
Abstract
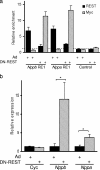
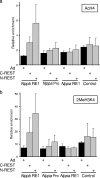
Cardiac hypertrophy is associated with a dramatic change in the gene expression profile of cardiac myocytes. Many genes important during development of the fetal heart but repressed in the adult tissue are reexpressed, resulting in gross physiological changes that lead to arrhythmias, cardiac failure, and sudden death. One transcription factor thought to be important in repressing the expression of fetal genes in the adult heart is the transcriptional repressor REST (repressor element 1-silencing transcription factor). Although REST has been shown to repress several fetal cardiac genes and inhibition of REST function is sufficient to induce cardiac hypertrophy, the molecular mechanisms employed in this repression are not known. Here we show that continued REST expression prevents increases in the levels of the BNP (Nppb) and ANP (Nppa) genes, encoding brain and atrial natriuretic peptides, in adult rat ventricular myocytes in response to endothelin-1 and that inhibition of REST results in increased expression of these genes in H9c2 cells. Increased expression of Nppb and Nppa correlates with increased histone H4 acetylation and histone H3 lysine 4 methylation of promoter-proximal regions of these genes. Furthermore, using deletions of individual REST repression domains, we show that the combined activities of two domains of REST are required to efficiently repress transcription of the Nppb gene; however, a single repression domain is sufficient to repress the Nppa gene. These data provide some of the first insights into the molecular mechanism that may be important for the changes in gene expression profile seen in cardiac hypertrophy.
Figures


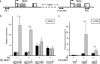
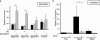

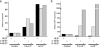
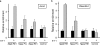
References
-
- Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. - PubMed
-
- Ballas, N., E. Battaglioli, F. Atouf, M. E. Andres, J. Chenoweth, M. E. Anderson, C. Burger, M. Moniwa, J. R. Davie, W. J. Bowers, H. J. Federoff, D. W. Rose, M. G. Rosenfeld, P. Brehm, and G. Mandel. 2001. Regulation of neuronal traits by a novel transcriptional complex. Neuron 31:353-365. - PubMed
-
- Ballas, N., C. Grunseich, D. D. Lu, J. C. Speh, and G. Mandel. 2005. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121:645-657. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
