The implications of alternative splicing in the ENCODE protein complement
- PMID: 17372197
- PMCID: PMC1838448
- DOI: 10.1073/pnas.0700800104
The implications of alternative splicing in the ENCODE protein complement
Abstract
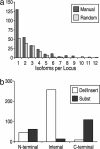
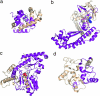
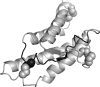
Alternative premessenger RNA splicing enables genes to generate more than one gene product. Splicing events that occur within protein coding regions have the potential to alter the biological function of the expressed protein and even to create new protein functions. Alternative splicing has been suggested as one explanation for the discrepancy between the number of human genes and functional complexity. Here, we carry out a detailed study of the alternatively spliced gene products annotated in the ENCODE pilot project. We find that alternative splicing in human genes is more frequent than has commonly been suggested, and we demonstrate that many of the potential alternative gene products will have markedly different structure and function from their constitutively spliced counterparts. For the vast majority of these alternative isoforms, little evidence exists to suggest they have a role as functional proteins, and it seems unlikely that the spectrum of conventional enzymatic or structural functions can be substantially extended through alternative splicing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
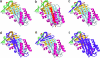
Similar articles
-
Structural genomics analysis of alternative splicing and application to isoform structure modeling.Proc Natl Acad Sci U S A. 2005 Dec 27;102(52):18920-5. doi: 10.1073/pnas.0506770102. Epub 2005 Dec 14. Proc Natl Acad Sci U S A. 2005. PMID: 16354838 Free PMC article.
-
Alternatively Spliced Homologous Exons Have Ancient Origins and Are Highly Expressed at the Protein Level.PLoS Comput Biol. 2015 Jun 10;11(6):e1004325. doi: 10.1371/journal.pcbi.1004325. eCollection 2015 Jun. PLoS Comput Biol. 2015. PMID: 26061177 Free PMC article.
-
Comparative proteomics reveals a significant bias toward alternative protein isoforms with conserved structure and function.Mol Biol Evol. 2012 Sep;29(9):2265-83. doi: 10.1093/molbev/mss100. Epub 2012 Mar 22. Mol Biol Evol. 2012. PMID: 22446687 Free PMC article.
-
Mechanisms and Regulation of Alternative Pre-mRNA Splicing.Annu Rev Biochem. 2015;84:291-323. doi: 10.1146/annurev-biochem-060614-034316. Epub 2015 Mar 12. Annu Rev Biochem. 2015. PMID: 25784052 Free PMC article. Review.
-
Function of alternative splicing.Gene. 2005 Jan 3;344:1-20. doi: 10.1016/j.gene.2004.10.022. Epub 2004 Dec 10. Gene. 2005. PMID: 15656968 Review.
Cited by
-
Alternative splicing in the regulation of cholesterol homeostasis.Curr Opin Lipidol. 2013 Apr;24(2):147-52. doi: 10.1097/MOL.0b013e32835cf284. Curr Opin Lipidol. 2013. PMID: 23314925 Free PMC article. Review.
-
FFPred 3: feature-based function prediction for all Gene Ontology domains.Sci Rep. 2016 Aug 26;6:31865. doi: 10.1038/srep31865. Sci Rep. 2016. PMID: 27561554 Free PMC article.
-
Pan-Tissue and -Cancer Analysis of ROR1 and ROR2 Transcript Variants Identify Novel Functional Significance for an Alternative Splice Variant of ROR1.Biomedicines. 2022 Oct 13;10(10):2559. doi: 10.3390/biomedicines10102559. Biomedicines. 2022. PMID: 36289823 Free PMC article.
-
Linear motifs confer functional diversity onto splice variants.Nucleic Acids Res. 2012 Aug;40(15):7123-31. doi: 10.1093/nar/gks442. Epub 2012 May 25. Nucleic Acids Res. 2012. PMID: 22638587 Free PMC article.
-
Association of TLR4 and TLR9 gene polymorphisms and haplotypes with cervicitis susceptibility.PLoS One. 2019 Jul 31;14(7):e0220330. doi: 10.1371/journal.pone.0220330. eCollection 2019. PLoS One. 2019. PMID: 31365550 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

