Genetic abnormalities involved in t(12;21) TEL-AML1 acute lymphoblastic leukemia: analysis by means of array-based comparative genomic hybridization
- PMID: 17374122
- PMCID: PMC11159317
- DOI: 10.1111/j.1349-7006.2007.00443.x
Genetic abnormalities involved in t(12;21) TEL-AML1 acute lymphoblastic leukemia: analysis by means of array-based comparative genomic hybridization
Abstract
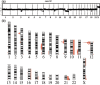
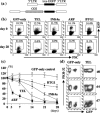
The TEL (ETV6)-AML1 (RUNX1) chimeric gene fusion is the most common genetic abnormality in childhood acute lymphoblastic leukemias. Evidence suggests that this chimeric gene fusion constitutes an initiating mutation that is necessary but insufficient for the development of leukemia. In a search for additional genetic events that could be linked to the development of leukemia, we applied a genome-wide array-comparative genomic hybridization technique to 24 TEL-AML1 leukemia samples and two cell lines. It was found that at least two chromosomal imbalances were involved in all samples. Recurrent regions of chromosomal imbalance (>10% of cases) and representative involved genes were gain of chromosomes 10 (17%) and 21q (25%; RUNX1) and loss of 12p13.2 (87%; TEL), 9p21.3 (29%; p16INK4a/ARF), 9p13.2 (25%; PAX5), 12q21.3 (25%; BTG1), 3p21 (21%; LIMD1), 6q21 (17%; AIM1 and BLIMP1), 4q31.23 (17%; NR3C2), 11q22-q23 (13%; ATM) and 19q13.11-q13.12 (13%; PDCD5). Enforced expression of TEL and to a lesser extent BTG1, both single genes known to be located in their respective minimum common region of loss, inhibited proliferation of the TEL-AML1 cell line Reh. Together, these findings suggest that some of the genes identified as lost by array-comparative genomic hybridization may partly account for the development of leukemia.
Figures




References
-
- Greaves MF, Maia AT, Wiemels JL, Ford AM. Leukemia in twins: lessons in natural history. Blood 2003; 102: 2321–33. - PubMed
-
- Zelent A, Greaves M, Enver T. Role of the TEL–AML1 fusion gene in the molecular pathogenesis of childhood acute lymphoblastic leukaemia. Oncogene 2004; 23: 4275–83. - PubMed
-
- Bernardin F, Yang Y, Cleaves R et al. TEL‐AML1, expressed from t(12;21) in human acute lymphocytic leukemia, induces acute leukemia in mice. Cancer Res 2002; 62: 3904–8. - PubMed
-
- Morrow M, Horton S, Kioussis D, Brady HJ, Williams O. TEL–AML1 promotes development of specific hematopoietic lineages consistent with preleukemic activity. Blood 2004; 103: 3890–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

