Isolation and functional characterization of a cDNA coding a hydroxycinnamoyltransferase involved in phenylpropanoid biosynthesis in Cynara cardunculus L
- PMID: 17374149
- PMCID: PMC1847684
- DOI: 10.1186/1471-2229-7-14
Isolation and functional characterization of a cDNA coding a hydroxycinnamoyltransferase involved in phenylpropanoid biosynthesis in Cynara cardunculus L
Abstract
Background: Cynara cardunculus L. is an edible plant of pharmaceutical interest, in particular with respect to the polyphenolic content of its leaves. It includes three taxa: globe artichoke, cultivated cardoon, and wild cardoon. The dominating phenolics are the di-caffeoylquinic acids (such as cynarin), which are largely restricted to Cynara species, along with their precursor, chlorogenic acid (CGA). The scope of this study is to better understand CGA synthesis in this plant.
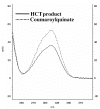
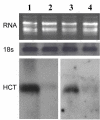
Results: A gene sequence encoding a hydroxycinnamoyltransferase (HCT) involved in the synthesis of CGA, was identified. Isolation of the gene sequence was achieved by using a PCR strategy with degenerated primers targeted to conserved regions of orthologous HCT sequences available. We have isolated a 717 bp cDNA which shares 84% aminoacid identity and 92% similarity with a tobacco gene responsible for the biosynthesis of CGA from p-coumaroyl-CoA and quinic acid. In silico studies revealed the globe artichoke HCT sequence clustering with one of the main acyltransferase groups (i.e. anthranilate N-hydroxycinnamoyl/benzoyltransferase). Heterologous expression of the full length HCT (GenBank accession DQ104740) cDNA in E. coli demonstrated that the recombinant enzyme efficiently synthesizes both chlorogenic acid and p-coumaroyl quinate from quinic acid and caffeoyl-CoA or p-coumaroyl-CoA, respectively, confirming its identity as a hydroxycinnamoyl-CoA: quinate HCT. Variable levels of HCT expression were shown among wild and cultivated forms of C. cardunculus subspecies. The level of expression was correlated with CGA content.
Conclusion: The data support the predicted involvement of the Cynara cardunculus HCT in the biosynthesis of CGA before and/or after the hydroxylation step of hydroxycinnamoyl esters.
Figures


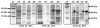
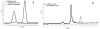


References
-
- Rottenberg A, Zohary D, Nevo E. Isozyme relationships between cultivated artichoke and the wild relatives. Genetic Resources and Crop Evolution. 1996;43:59–62.
-
- Lanteri S, Saba E, Cadinu M, Mallica GM, Baghino L, Portis E. Amplified fragment length polymorphism for genetic diversity assessment in globe artichoke. Theor Appl Genet. 2004;108:1534–1544. - PubMed
-
- Acquadro A, Portis E, Lee D, Donini P, Lanteri S. Development and characterisation of microsatellite markers in Cynara cardunculus L. Genome. 2005;48:217–225. - PubMed
-
- Dogan S, Turan Y, Erturk H, Arslan O. Characterization and purification of polyphenol oxidase from Artichoke (Cynara scolymus L.) J Agric Food Chem. 2005;53:776–785. - PubMed
-
- Slanina J, Taborska E, Musil P. Determination of cynarine in the decoctions of the artichoke (Cynara cardunculus L.) by the HPLC method. Cesko-SloV Farm. 1993;42:265–268.
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

