In-Silico docking of HIV-1 integrase inhibitors reveals a novel drug type acting on an enzyme/DNA reaction intermediate
- PMID: 17374162
- PMCID: PMC1847836
- DOI: 10.1186/1742-4690-4-21
In-Silico docking of HIV-1 integrase inhibitors reveals a novel drug type acting on an enzyme/DNA reaction intermediate
Abstract
Background: HIV-1 integrase (IN) is an emerging drug target, as IN strand transfer inhibitors (INSTIs) are proving potent antiretroviral agents in clinical trials. One credible theory sees INSTIs as docking at the cellular (acceptor) DNA-binding site after IN forms a transitional complex with viral (donor) DNA. However, mapping of the DNA and INSTI binding sites within the IN catalytic core domain (CCD) has been uncertain.
Methods: Structural superimpositions were conducted using the SWISS PDB and Cn3D free software. Docking simulations of INSTIs were run by a widely validated genetic algorithm (GOLD).
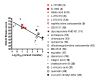
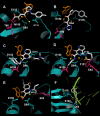
Results: Structural superimpositions suggested that a two-metal model for HIV-1 IN CCD in complex with small molecule, 1-(5-chloroindol-3-yl)-3-(tetrazoyl)-1,3-propandione-ene (5CITEP) could be used as a surrogate for an IN/viral DNA complex, because it allowed replication of contacts documented biochemically in viral DNA/IN complexes or displayed by a crystal structure of the IN-related enzyme Tn5 transposase in complex with transposable DNA. Docking simulations showed that the fitness of different compounds for the catalytic cavity of the IN/5CITEP complex significantly (P < 0.01) correlated with their 50% inhibitory concentrations (IC50s) in strand transfer assays in vitro. The amino acids involved in inhibitor binding matched those involved in drug resistance. Both metal binding and occupation of the putative viral DNA binding site by 5CITEP appeared to be important for optimal drug/ligand interactions. The docking site of INSTIs appeared to overlap with a putative acceptor DNA binding region adjacent to but distinct from the putative donor DNA binding site, and homologous to the nucleic acid binding site of RNAse H. Of note, some INSTIs such as 4,5-dihydroxypyrimidine carboxamides/N-Alkyl-5-hydroxypyrimidinone carboxamides, a highly promising drug class including raltegravir/MK-0518 (now in clinical trials), displayed interactions with IN reminiscent of those displayed by fungal molecules from Fusarium sp., shown in the 1990s to inhibit HIV-1 integration.
Conclusion: The 3D model presented here supports the idea that INSTIs dock at the putative acceptor DNA-binding site in a IN/viral DNA complex. This mechanism of enzyme inhibition, likely to be exploited by some natural products, might disclose future strategies for inhibition of nucleic acid-manipulating enzymes.
Figures

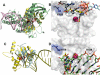
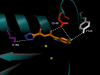

References
-
- DeJesus E, Berger D, Markowitz M, Cohen C, Hawkins T, Ruane P, Elion R, Farthing C, Zhong L, Cheng AK, McColl D, Kearney BP, for the 183-0101 Study Team Antiviral activity, pharmacokinetics, and dose response of the HIV-1 integrase inhibitor GS-9137 (JTK-303) in treatment-naive and treatment-experienced patients. J Acquir Immune Defic Syndr. 2006;1:1–5. - PubMed
-
- Markowitz M, Morales-Ramirez JO, Nguyen BY, Kovacs CM, Steigbigel RT, Cooper DA, Liporace R, Schwartz R, Isaacs R, Gilde LR, Wenning L, Zhao J, Teppler H, the Protocol 004 Study Team Antiretroviral Activity, Pharmacokinetics, and Tolerability of MK-0518 a Novel Inhibitor of HIV-1 Integrase, Dosed As Monotherapy for 10 Days in Treatment-Naive HIV-1-Infected Individuals. J Acquir Immune Defic Syndr. 2006;5:509–515. 2006 Dec 15. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

