BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis
- PMID: 17376028
- PMCID: PMC1925254
- DOI: 10.1042/BJ20070102
BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis
Abstract
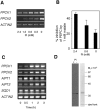
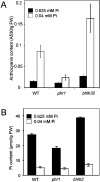
P(i) (inorganic phosphate) limitation severely impairs plant growth and reduces crop yield. Hence plants have evolved several biochemical and morphological responses to P(i) starvation that both enhance uptake and conserve use. The mechanisms involved in P(i) sensing and signal transduction are not completely understood. In the present study we report that a previously uncharacterized transcription factor, BHLH32, acts as a negative regulator of a range of P(i) starvation-induced processes in Arabidopsis. In bhlh32 mutant plants in P(i)-sufficient conditions, expression of several P(i) starvation-induced genes, formation of anthocyanins, total P(i) content and root hair formation were all significantly increased compared with the wild-type. Among the genes negatively regulated by BHLH32 are those encoding PPCK (phosphoenolpyruvate carboxylase kinase), which is involved in modifying metabolism so that P(i) is spared. The present study has shown that PPCK genes are rapidly induced by P(i) starvation leading to increased phosphorylation of phosphoenolpyruvate carboxylase. Furthermore, several Arabidopsis proteins that regulate epidermal cell differentiation [TTG1 (TRANSPARENT TESTA GLABRA1), GL3 (GLABRA3) and EGL3 (ENHANCER OF GL3)] positively regulate PPCK gene expression in response to P(i) starvation. BHLH32 can physically interact with TTG1 and GL3. We propose that BHLH32 interferes with the function of TTG1-containing complexes and thereby affects several biochemical and morphological processes that respond to P(i) availability.
Figures




References
-
- Raghothama K. G. Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:665–693. - PubMed
-
- Bates T. R., Lynch J. P. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 1996;19:529–538.
-
- Vance C. P., Uhde-Stone C., Allan D. L. Phosphorus acquisition and use: critical adaptations by plants for securing a non-renewable resource. New Phytol. 2003;157:423–447. - PubMed
-
- Ticconi C. A., Abel S. Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci. 2004;9:548–555. - PubMed
-
- Hammond J. P., Bennett M. J., Bowen H. C., Broadley M. R., Eastwood D. C., May S. T., Rahn C., Swarup R., Woolaway K. E., White P. J. Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 2003;132:578–596. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

