Temperature-dependent RNP conformational rearrangements: analysis of binary complexes of primary binding proteins with 16 S rRNA
- PMID: 17376481
- PMCID: PMC2265208
- DOI: 10.1016/j.jmb.2007.02.064
Temperature-dependent RNP conformational rearrangements: analysis of binary complexes of primary binding proteins with 16 S rRNA
Abstract
Ribonucleoprotein particles (RNPs) are important components of all living systems, and the assembly of these particles is an intricate, often multistep, process. The 30 S ribosomal subunit is composed of one large RNA (16 S rRNA) and 21 ribosomal proteins (r-proteins). In vitro studies have revealed that assembly of the 30 S subunit is a temperature-dependent process involving sequential binding of r-proteins and conformational changes of 16 S rRNA. Additionally, a temperature-dependent conformational rearrangement was reported for a complex of primary r-protein S4 and 16 S rRNA. Given these observations, a systematic study of the temperature-dependence of 16 S rRNA architecture in individual complexes with the other five primary binding proteins (S7, S8, S15, S17, and S20) was performed. While all primary binding r-proteins bind 16 S rRNA at low temperature, not all r-proteins/16 S rRNA complexes undergo temperature-dependent conformational rearrangements. Some RNPs achieve the same conformation regardless of temperature, others show minor adjustments in 16 S rRNA conformation upon heating and, finally, others undergo significant temperature-dependent changes. Some of the architectures achieved in these rearrangements are consistent with subsequent downstream assembly events such as assembly of the secondary and tertiary binding r-proteins. The differential interaction of 16 S rRNA with r-proteins illustrates a means for controlling the sequential assembly pathway for complex RNPs and may offer insights into aspects of RNP assembly in general.
Figures

Modified in vitro 30S subunit assembly map. The 16S rRNA is represented by a rectangle in a 5' to 3' direction. The arrows indicate the co-dependencies for the assembly of the r-proteins. The size of the arrow indicates the relative strength of the assembly dependency between components. The r-proteins shown in the white region are primary binding r-proteins. The r-proteins shown in white in the light gray and dark gray box indicate secondary, and tertiary binding r-proteins, respectively. S6 and S18 are enclosed in a box to indicate that they bind as a heterodimer.
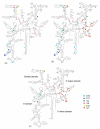
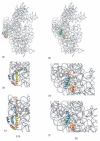
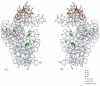
Crystal structure of the 16S rRNA from the E. coli 30S subunit with all the primary proteins. The 16S rRNA is shown in gray, and the r-proteins are S4 green, S7 red, S8 magenta, S15 bright yellow, S17 dark purple and S20 light blue, as in Figure 1a. The 3-D parts of the 30S subunit are indicated while the corresponding domains from the 16S rRNA secondary structure are specified in paranthesis. All the Figures containing 3-D structures were prepared using Pymol, and the pdb file 2AW7.





Similar articles
-
Protein-induced conformational changes in 16 S ribosomal RNA during the initial assembly steps of the Escherichia coli 30 S ribosomal subunit.J Mol Biol. 1989 Nov 20;210(2):323-36. doi: 10.1016/0022-2836(89)90334-3. J Mol Biol. 1989. PMID: 2689654
-
Assembly of the central domain of the 30S ribosomal subunit: roles for the primary binding ribosomal proteins S15 and S8.J Mol Biol. 2003 Jul 4;330(2):373-83. doi: 10.1016/s0022-2836(03)00586-2. J Mol Biol. 2003. PMID: 12823975
-
Ribosomal protein-dependent orientation of the 16 S rRNA environment of S15.J Mol Biol. 2004 Jan 30;335(5):1173-85. doi: 10.1016/j.jmb.2003.11.031. J Mol Biol. 2004. PMID: 14729335
-
RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA.Science. 1989 May 19;244(4906):783-90. doi: 10.1126/science.2658053. Science. 1989. PMID: 2658053 Review.
-
Structure and dynamics of ribosomal RNA.Curr Opin Struct Biol. 1998 Jun;8(3):294-300. doi: 10.1016/s0959-440x(98)80061-4. Curr Opin Struct Biol. 1998. PMID: 9666324 Review.
Cited by
-
RNA folding and ribosome assembly.Curr Opin Chem Biol. 2008 Dec;12(6):667-73. doi: 10.1016/j.cbpa.2008.09.024. Epub 2008 Oct 18. Curr Opin Chem Biol. 2008. PMID: 18935976 Free PMC article. Review.
-
A new layer of rRNA regulation by small interference RNAs and the nuclear RNAi pathway.RNA Biol. 2017 Nov 2;14(11):1492-1498. doi: 10.1080/15476286.2017.1341034. Epub 2017 Jul 21. RNA Biol. 2017. PMID: 28640690 Free PMC article. Review.
-
Proteomic Response to Rising Temperature in the Marine Cyanobacterium Synechococcus Grown in Different Nitrogen Sources.Front Microbiol. 2019 Aug 23;10:1976. doi: 10.3389/fmicb.2019.01976. eCollection 2019. Front Microbiol. 2019. PMID: 31507578 Free PMC article.
-
Assembly of the 5' and 3' minor domains of 16S ribosomal RNA as monitored by tethered probing from ribosomal protein S20.J Mol Biol. 2008 Feb 8;376(1):92-108. doi: 10.1016/j.jmb.2007.10.083. Epub 2007 Nov 6. J Mol Biol. 2008. PMID: 18155048 Free PMC article.
-
Global stabilization of rRNA structure by ribosomal proteins S4, S17, and S20.J Mol Biol. 2009 Sep 25;392(3):666-77. doi: 10.1016/j.jmb.2009.07.032. Epub 2009 Jul 16. J Mol Biol. 2009. PMID: 19616559 Free PMC article.
References
-
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–34. - PubMed
-
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–8. - PubMed
-
- Wimberly BT, Brodersen DE, Clemons WM, Jr., Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–39. - PubMed
-
- Hosaka H, Nakagawa A, Tanaka I, Harada N, Sano K, Kimura M, Yao M, Wakatsuki S. Ribosomal protein S7: a new RNA-binding motif with structural similarities to a DNA architectural factor. Structure. 1997;5:1199–1208. - PubMed
-
- Wimberly BT, White SW, Ramakrishnan V. The structure of ribosomal protein S7 at 1.9 A resolution reveals a beta-hairpin motif that binds double-stranded nucleic acids. Structure. 1997;5:1187–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

