Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes
- PMID: 17376915
- PMCID: PMC1900311
- DOI: 10.1128/JVI.01714-06
Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes
Abstract
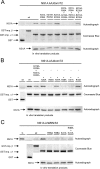
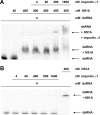
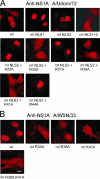
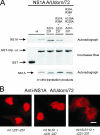
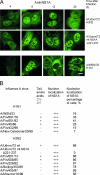
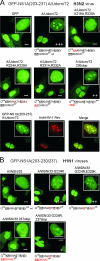
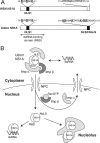
Influenza A virus nonstructural protein 1 (NS1A protein) is a virulence factor which is targeted into the nucleus. It is a multifunctional protein that inhibits host cell pre-mRNA processing and counteracts host cell antiviral responses. We show that the NS1A protein can interact with all six human importin alpha isoforms, indicating that the nuclear translocation of NS1A protein is mediated by the classical importin alpha/beta pathway. The NS1A protein of the H1N1 (WSN/33) virus has only one N-terminal arginine- or lysine-rich nuclear localization signal (NLS1), whereas the NS1A protein of the H3N2 subtype (Udorn/72) virus also has a second C-terminal NLS (NLS2). NLS1 is mapped to residues 35 to 41, which also function in the double-stranded RNA-binding activity of the NS1A protein. NLS2 was created by a 7-amino-acid C-terminal extension (residues 231 to 237) that became prevalent among human influenza A virus types isolated between the years 1950 to 1987. NLS2 includes basic amino acids at positions 219, 220, 224, 229, 231, and 232. Surprisingly, NLS2 also forms a functional nucleolar localization signal NoLS, a function that was retained in H3N2 type virus NS1A proteins even without the C-terminal extension. It is likely that the evolutionarily well-conserved nucleolar targeting function of NS1A protein plays a role in the pathogenesis of influenza A virus.
Figures


References
-
- Belmont, A. 2003. Dynamics of chromatin, proteins, and bodies within the cell nucleus. Curr. Opin. Cell Biol. 15:304-310. - PubMed
-
- Birbach, A., S. T. Bailey, S. Ghosh, and J. A. Schmid. 2004. Cytosolic, nuclear and nucleolar localization signals determine subcellular distribution and activity of the NF-κB inducing kinase NIK. J. Cell Sci. 117:3615-3624. - PubMed
-
- Bornholdt, Z. A., and B. V. Prasad. 2006. X-ray structure of influenza virus NS1 effector domain. Nat. Struct. Mol. Biol. 13:559-560. - PubMed
-
- Carmo-Fonseca, M. 2002. The contribution of nuclear compartmentalization to gene regulation. Cell 108:513-521. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

