Rotavirus NSP4 interacts with both the amino- and carboxyl-termini of caveolin-1
- PMID: 17379346
- PMCID: PMC1978065
- DOI: 10.1016/j.virusres.2007.02.004
Rotavirus NSP4 interacts with both the amino- and carboxyl-termini of caveolin-1
Abstract
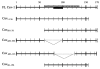
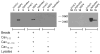
Rotavirus NSP4 plays multiple roles in viral pathogenesis, morphogenesis and replication. We previously reported a direct interaction between full-length NSP4 and the enterotoxic peptide composed of NSP4 residues 114-135 with full-length caveolin-1, the structural protein of caveolae. Caveolin-1 forms a hairpin loop in the cytoplasmic leaflet of plasma membrane caveolae. This unique orientation results in both termini of caveolin-1 exposed to the cytoplasm. The goal of this study was to map the caveolin-1 residues that interact with NSP4 to obtain a more complete picture of this binding event. Utilizing reverse yeast two-hybrid analyses and direct peptide binding assays, the NSP4 binding site was localized to caveolin-1 residues 2-22 and 161-178, at the amino- and carboxyl-termini, respectively. However, NSP4 binding to one of the termini was sufficient for the interaction.
Figures



Similar articles
-
The rotavirus enterotoxin NSP4 directly interacts with the caveolar structural protein caveolin-1.J Virol. 2006 Mar;80(6):2842-54. doi: 10.1128/JVI.80.6.2842-2854.2006. J Virol. 2006. PMID: 16501093 Free PMC article.
-
Mutational analysis of the rotavirus NSP4 enterotoxic domain that binds to caveolin-1.Virol J. 2013 Nov 13;10:336. doi: 10.1186/1743-422X-10-336. Virol J. 2013. PMID: 24220211 Free PMC article.
-
Comparison of NSP4 protein between group A and B human rotaviruses: detection of novel diarrhea-causing sequences in group B NSP4.Arch Virol. 2006 Jan;151(1):173-82. doi: 10.1007/s00705-005-0616-8. Epub 2005 Aug 23. Arch Virol. 2006. PMID: 16132179
-
Rotavirus NSP4: a multifunctional viral enterotoxin.Viral Immunol. 2005;18(1):27-40. doi: 10.1089/vim.2005.18.27. Viral Immunol. 2005. PMID: 15802952 Review. No abstract available.
-
Rotavirus protein expression is important for virus assembly and pathogenesis.Arch Virol Suppl. 1996;12:69-77. doi: 10.1007/978-3-7091-6553-9_8. Arch Virol Suppl. 1996. PMID: 9015103 Review.
Cited by
-
Multifaceted Functions of Host Cell Caveolae/Caveolin-1 in Virus Infections.Viruses. 2020 Apr 26;12(5):487. doi: 10.3390/v12050487. Viruses. 2020. PMID: 32357558 Free PMC article. Review.
-
Caveolae as Potential Hijackable Gates in Cell Communication.Front Cell Dev Biol. 2020 Oct 27;8:581732. doi: 10.3389/fcell.2020.581732. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33195223 Free PMC article. Review.
-
Novel pentameric structure of the diarrhea-inducing region of the rotavirus enterotoxigenic protein NSP4.J Virol. 2011 Dec;85(23):12721-32. doi: 10.1128/JVI.00349-11. Epub 2011 Sep 14. J Virol. 2011. PMID: 21917949 Free PMC article.
-
A new N-terminal recognition domain in caveolin-1 interacts with sterol carrier protein-2 (SCP-2).Biochemistry. 2007 Jul 17;46(28):8301-14. doi: 10.1021/bi7002636. Epub 2007 Jun 20. Biochemistry. 2007. PMID: 17580960 Free PMC article.
-
The Dengue Virus Nonstructural Protein 1 (NS1) Is Secreted from Mosquito Cells in Association with the Intracellular Cholesterol Transporter Chaperone Caveolin Complex.J Virol. 2019 Feb 5;93(4):e01985-18. doi: 10.1128/JVI.01985-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30463973 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

