Minding the gap: the underground functions of BRCA1 and BRCA2 at stalled replication forks
- PMID: 17379580
- PMCID: PMC2989184
- DOI: 10.1016/j.dnarep.2007.02.020
Minding the gap: the underground functions of BRCA1 and BRCA2 at stalled replication forks
Abstract
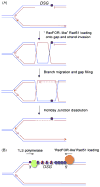
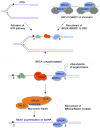
The hereditary breast and ovarian cancer predisposition genes, BRCA1 and BRCA2, participate in the repair of DNA double strand breaks by homologous recombination. Circumstantial evidence implicates these genes in recombinational responses to DNA polymerase stalling during the S phase of the cell cycle. These responses play a key role in preventing genomic instability and cancer. Here, we review the current literature implicating the BRCA pathway in HR at stalled replication forks and explore the hypothesis that BRCA1 and BRCA2 participate in the recombinational resolution of single stranded DNA lesions termed "daughter strand gaps", generated during replication across a damaged DNA template.
Figures


Similar articles
-
BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks.Nature. 2014 Jun 26;510(7506):556-9. doi: 10.1038/nature13295. Epub 2014 Apr 28. Nature. 2014. PMID: 24776801 Free PMC article.
-
Revisiting the BRCA-pathway through the lens of replication gap suppression: "Gaps determine therapy response in BRCA mutant cancer".DNA Repair (Amst). 2021 Nov;107:103209. doi: 10.1016/j.dnarep.2021.103209. Epub 2021 Aug 13. DNA Repair (Amst). 2021. PMID: 34419699 Free PMC article. Review.
-
Therapeutic exploitation of tumor cell defects in homologous recombination.Anticancer Agents Med Chem. 2008 May;8(4):448-60. doi: 10.2174/187152008784220267. Anticancer Agents Med Chem. 2008. PMID: 18473729 Review.
-
BRCA1 and BRCA2 protect against oxidative DNA damage converted into double-strand breaks during DNA replication.DNA Repair (Amst). 2015 Jun;30:11-20. doi: 10.1016/j.dnarep.2015.03.002. Epub 2015 Mar 17. DNA Repair (Amst). 2015. PMID: 25836596 Free PMC article.
-
Targeting the DNA repair defect of BRCA tumours.Curr Opin Pharmacol. 2005 Aug;5(4):388-93. doi: 10.1016/j.coph.2005.03.006. Curr Opin Pharmacol. 2005. PMID: 15955736 Review.
Cited by
-
S100A11 plays a role in homologous recombination and genome maintenance by influencing the persistence of RAD51 in DNA repair foci.Cell Cycle. 2016 Oct 17;15(20):2766-79. doi: 10.1080/15384101.2016.1220457. Epub 2016 Aug 11. Cell Cycle. 2016. PMID: 27590262 Free PMC article.
-
GEMIN2 promotes accumulation of RAD51 at double-strand breaks in homologous recombination.Nucleic Acids Res. 2010 Aug;38(15):5059-74. doi: 10.1093/nar/gkq271. Epub 2010 Apr 19. Nucleic Acids Res. 2010. PMID: 20403813 Free PMC article.
-
Genetic evidence for single-strand lesions initiating Nbs1-dependent homologous recombination in diversification of Ig v in chicken B lymphocytes.PLoS Genet. 2009 Jan;5(1):e1000356. doi: 10.1371/journal.pgen.1000356. Epub 2009 Jan 30. PLoS Genet. 2009. PMID: 19180185 Free PMC article.
-
BRCA1 is required for postreplication repair after UV-induced DNA damage.Mol Cell. 2011 Oct 21;44(2):235-51. doi: 10.1016/j.molcel.2011.09.002. Epub 2011 Sep 29. Mol Cell. 2011. PMID: 21963239 Free PMC article.
-
RNF20-mediated H2B monoubiquitination protects stalled forks from degradation and promotes fork restart.EMBO Rep. 2025 Aug;26(15):3773-3803. doi: 10.1038/s44319-025-00497-3. Epub 2025 Jun 10. EMBO Rep. 2025. PMID: 40495033 Free PMC article.
References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. - PubMed
-
- Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25:156–165. - PubMed
-
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

