Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression
- PMID: 17380126
- PMCID: PMC1852780
- DOI: 10.1038/sj.emboj.7601654
Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression
Abstract
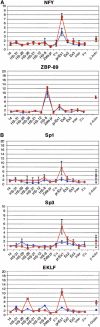
To understand how mammalian genes are regulated from their natural chromosomal environment, we have analysed the molecular events occurring throughout a 150 kb chromatin segment containing the alpha globin gene locus as it changes from a poised, silent state in erythroid progenitors, to the fully activated state in late, erythroid cells. Active transcription requires the late recruitment of general transcription factors, mediator and Pol II not only to the promoter but also to its remote regulatory elements. Natural mutants of the alpha cluster show that whereas recruitment of the pre-initiation complex to the upstream elements occurs independently, recruitment to the promoter is largely dependent on the regulatory elements. An improved, quantitative chromosome conformation capture analysis demonstrates that this recruitment is associated with a conformational change, in vivo, apposing the promoter with its remote regulators, consistent with a chromosome looping mechanism. These findings point to a general mechanism by which a gene can be held in a poised state until the appropriate stage for expression, coordinating the level and timing of gene expression during terminal differentiation.
Figures




References
-
- Bartlett J, Blagojevic J, Carter D, Eskiw C, Fromaget M, Job C, Shamsher M, Trindade IF, Xu M, Cook PR (2006) Specialized transcription factories. Biochem Soc Symp 73: 67–75 - PubMed
-
- Bender MA, Mehaffey MG, Telling A, Hug B, Ley TJ, Groudine M, Fiering S (2000) Independent formation of DnaseI hypersensitive sites in the murine beta-globin locus control region. Blood 95: 3600–3604 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

