RNA channelling by the archaeal exosome
- PMID: 17380186
- PMCID: PMC1866195
- DOI: 10.1038/sj.embor.7400945
RNA channelling by the archaeal exosome
Abstract
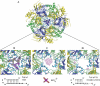
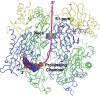
Exosomes are complexes containing 3' --> 5' exoribonucleases that have important roles in processing, decay and quality control of various RNA molecules. Archaeal exosomes consist of a hexameric core of three active RNase PH subunits (ribosomal RNA processing factor (Rrp)41) and three inactive RNase PH subunits (Rrp42). A trimeric ring of subunits with putative RNA-binding domains (Rrp4/cep1 synthetic lethality (Csl)4) is positioned on top of the hexamer on the opposite side to the RNA degrading sites. Here, we present the 1.6 A resolution crystal structure of the nine-subunit exosome of Sulfolobus solfataricus and the 2.3 A structure of this complex bound to an RNA substrate designed to be partly trimmed rather than completely degraded. The RNA binds both at the active site on one side of the molecule and on the opposite side in the narrowest constriction of the central channel. Multiple substrate-binding sites and the entrapment of the substrate in the central channel provide a rationale for the processive degradation of extended RNAs and the stalling of structured RNAs.
Figures


Comment in
-
The exosome, plugged.EMBO Rep. 2007 May;8(5):456-7. doi: 10.1038/sj.embor.7400961. EMBO Rep. 2007. PMID: 17471261 Free PMC article. No abstract available.
Similar articles
-
Structural basis of 3' end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core.Mol Cell. 2005 Nov 11;20(3):473-81. doi: 10.1016/j.molcel.2005.10.020. Mol Cell. 2005. PMID: 16285928
-
The archaeal exosome core is a hexameric ring structure with three catalytic subunits.Nat Struct Mol Biol. 2005 Jul;12(7):575-81. doi: 10.1038/nsmb952. Epub 2005 Jun 12. Nat Struct Mol Biol. 2005. PMID: 15951817
-
Rrp4 and Csl4 are needed for efficient degradation but not for polyadenylation of synthetic and natural RNA by the archaeal exosome.Biochemistry. 2008 Dec 16;47(50):13158-68. doi: 10.1021/bi8012214. Biochemistry. 2008. PMID: 19053279
-
Structure and function of the archaeal exosome.Wiley Interdiscip Rev RNA. 2014 Sep-Oct;5(5):623-35. doi: 10.1002/wrna.1234. Epub 2014 Apr 30. Wiley Interdiscip Rev RNA. 2014. PMID: 24789718 Review.
-
The archaeal exosome.Adv Exp Med Biol. 2010;702:29-38. Adv Exp Med Biol. 2010. PMID: 21618872 Review.
Cited by
-
Insight into the RNA Exosome Complex Through Modeling Pontocerebellar Hypoplasia Type 1b Disease Mutations in Yeast.Genetics. 2017 Jan;205(1):221-237. doi: 10.1534/genetics.116.195917. Epub 2016 Oct 24. Genetics. 2017. PMID: 27777260 Free PMC article.
-
Crystal structure of human polynucleotide phosphorylase: insights into its domain function in RNA binding and degradation.Nucleic Acids Res. 2012 May;40(9):4146-57. doi: 10.1093/nar/gkr1281. Epub 2011 Dec 30. Nucleic Acids Res. 2012. PMID: 22210891 Free PMC article.
-
Structural components and architectures of RNA exosomes.Adv Exp Med Biol. 2010;702:9-28. Adv Exp Med Biol. 2010. PMID: 21618871 Free PMC article. Review.
-
Reining in RNA. Workshop on intracellular RNA localization and localized translation.EMBO Rep. 2008 Jan;9(1):22-6. doi: 10.1038/sj.embor.7401140. Epub 2007 Dec 14. EMBO Rep. 2008. PMID: 18084187 Free PMC article. No abstract available.
-
Subcellular localization of RNA degrading proteins and protein complexes in prokaryotes.RNA Biol. 2011 Jan-Feb;8(1):49-54. doi: 10.4161/rna.8.1.14066. Epub 2011 Jan 1. RNA Biol. 2011. PMID: 21289488 Free PMC article.
References
-
- Bousquet-Antonelli C, Presutti C, Tollervey D (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 - PubMed
-
- Briggs MW, Burkard KT, Butler JS (1998) Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8S rRNA 3′ end formation. J Biol Chem 273: 13255–13263 - PubMed
-
- Buttner K, Wenig K, Hopfner KP (2005) Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell 20: 461–471 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

