Autistic-like phenotypes in Cadps2-knockout mice and aberrant CADPS2 splicing in autistic patients
- PMID: 17380209
- PMCID: PMC1821065
- DOI: 10.1172/JCI29031
Autistic-like phenotypes in Cadps2-knockout mice and aberrant CADPS2 splicing in autistic patients
Abstract
Autism, characterized by profound impairment in social interactions and communicative skills, is the most common neurodevelopmental disorder, and its underlying molecular mechanisms remain unknown. Ca(2+)-dependent activator protein for secretion 2 (CADPS2; also known as CAPS2) mediates the exocytosis of dense-core vesicles, and the human CADPS2 is located within the autism susceptibility locus 1 on chromosome 7q. Here we show that Cadps2-knockout mice not only have impaired brain-derived neurotrophic factor release but also show autistic-like cellular and behavioral phenotypes. Moreover, we found an aberrant alternatively spliced CADPS2 mRNA that lacks exon 3 in some autistic patients. Exon 3 was shown to encode the dynactin 1-binding domain and affect axonal CADPS2 protein distribution. Our results suggest that a disturbance in CADPS2-mediated neurotrophin release contributes to autism susceptibility.
Figures




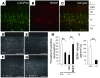
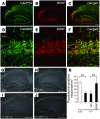

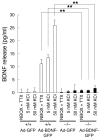




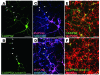
Comment in
-
Comment on "Autistic-like phenotypes in Cadps2-knockout mice and aberrant CADPS2 splicing in autistic patients".J Clin Invest. 2009 Apr;119(4):679-80; author reply 680-1. doi: 10.1172/JCI38620. Epub 2009 Apr 1. J Clin Invest. 2009. PMID: 19339759 Free PMC article. No abstract available.
Similar articles
-
Ca(2+)-dependent activator protein for secretion 2 and autistic-like phenotypes.Neurosci Res. 2010 Jul;67(3):197-202. doi: 10.1016/j.neures.2010.03.006. Epub 2010 Mar 17. Neurosci Res. 2010. PMID: 20302894 Review.
-
Comment on "Autistic-like phenotypes in Cadps2-knockout mice and aberrant CADPS2 splicing in autistic patients".J Clin Invest. 2009 Apr;119(4):679-80; author reply 680-1. doi: 10.1172/JCI38620. Epub 2009 Apr 1. J Clin Invest. 2009. PMID: 19339759 Free PMC article. No abstract available.
-
Alternative splicing variations in mouse CAPS2: differential expression and functional properties of splicing variants.BMC Neurosci. 2007 Apr 12;8:25. doi: 10.1186/1471-2202-8-25. BMC Neurosci. 2007. PMID: 17428348 Free PMC article.
-
Autistic-like behavioral phenotypes in a mouse model with copy number variation of the CAPS2/CADPS2 gene.FEBS Lett. 2013 Jan 4;587(1):54-9. doi: 10.1016/j.febslet.2012.10.047. Epub 2012 Nov 15. FEBS Lett. 2013. PMID: 23159942
-
Developmentally regulated Ca2+-dependent activator protein for secretion 2 (CAPS2) is involved in BDNF secretion and is associated with autism susceptibility.Cerebellum. 2009 Sep;8(3):312-22. doi: 10.1007/s12311-009-0097-5. Epub 2009 Feb 24. Cerebellum. 2009. PMID: 19238500 Review.
Cited by
-
Alterations of presynaptic proteins in autism spectrum disorder.Front Mol Neurosci. 2022 Nov 17;15:1062878. doi: 10.3389/fnmol.2022.1062878. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36466804 Free PMC article. Review.
-
Chd7 is indispensable for mammalian brain development through activation of a neuronal differentiation programme.Nat Commun. 2017 Mar 20;8:14758. doi: 10.1038/ncomms14758. Nat Commun. 2017. PMID: 28317875 Free PMC article.
-
Role of Imaging Genetics in Alzheimer's Disease: A Systematic Review and Current Update.CNS Neurol Disord Drug Targets. 2024;23(9):1143-1156. doi: 10.2174/0118715273264879231027070642. CNS Neurol Disord Drug Targets. 2024. PMID: 38243986
-
The molecular basis of cognitive deficits in pervasive developmental disorders.Learn Mem. 2012 Aug 16;19(9):434-43. doi: 10.1101/lm.025007.111. Learn Mem. 2012. PMID: 22904374 Free PMC article. Review.
-
CADPS functional mutations in patients with bipolar disorder increase the sensitivity to stress.Mol Psychiatry. 2022 Feb;27(2):1145-1157. doi: 10.1038/s41380-021-01151-9. Epub 2022 Feb 15. Mol Psychiatry. 2022. PMID: 35169262
References
-
- World Health Organization. 1992. The ICD-10 classification of mental and behavioural disorders: clinical descriptions and diagnostic guidelines. World Health Organization. Geneva, Switzerland. http://www.who.int/substance_abuse/terminology/icd_10/en/index.html.
-
- American Psychiatric Association. 1994. Diagnostic and statistical manual of mental disorders: DSM-IV. American Psychiatric Publishing Inc. Washington, DC, USA. 886 pp.
-
- Muhle R., Trentacoste S.V., Rapin I. The genetics of autism. Pediatrics. 2004;113:e472–e486. - PubMed
-
- Folstein S.E., Rosen-Sheidley B. Genetics of autism: complex aetiology for a heterogeneous disorder. Nat. Rev. Genet. 2001;2:943–955. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

