Structural biology of RNA silencing and its functional implications
- PMID: 17381284
- PMCID: PMC4689314
- DOI: 10.1101/sqb.2006.71.053
Structural biology of RNA silencing and its functional implications
Abstract
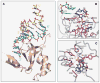
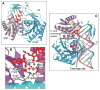
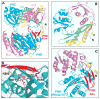
We outline structure-function contributions from our laboratories on protein-RNA recognition events that monitor siRNA length, 5 -phosphate and 2-nucleotide 3 overhangs, as well as the architecture of Argonaute, its externally bound siRNA complex, and Argonaute-based models involving guide-strand-mediated mRNA binding, cleavage, and release.
Figures



Similar articles
-
The Argonautes.Cold Spring Harb Symp Quant Biol. 2006;71:67-72. doi: 10.1101/sqb.2006.71.048. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381282 Review.
-
Molecular biology. Argonaute journeys into the heart of RISC.Science. 2004 Sep 3;305(5689):1409-10. doi: 10.1126/science.1103076. Science. 2004. PMID: 15353786 No abstract available.
-
Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein.Nature. 2005 Mar 31;434(7033):666-70. doi: 10.1038/nature03514. Nature. 2005. PMID: 15800629 Free PMC article.
-
Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex.Nature. 2005 Mar 31;434(7033):663-6. doi: 10.1038/nature03462. Nature. 2005. PMID: 15800628 Free PMC article.
-
Argonaute proteins: mediators of RNA silencing.Mol Cell. 2007 Jun 8;26(5):611-23. doi: 10.1016/j.molcel.2007.05.001. Mol Cell. 2007. PMID: 17560368 Review.
Cited by
-
Knocking down Insulin Receptor in Pancreatic Beta Cell lines with Lentiviral-Small Hairpin RNA Reduces Glucose-Stimulated Insulin Secretion via Decreasing the Gene Expression of Insulin, GLUT2 and Pdx1.Int J Mol Sci. 2018 Mar 26;19(4):985. doi: 10.3390/ijms19040985. Int J Mol Sci. 2018. PMID: 29587416 Free PMC article.
-
Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex.Nature. 2008 Dec 18;456(7224):921-6. doi: 10.1038/nature07666. Nature. 2008. PMID: 19092929 Free PMC article.
-
Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes.Nature. 2009 Oct 8;461(7265):754-61. doi: 10.1038/nature08434. Nature. 2009. PMID: 19812667 Free PMC article.
-
Structural basis for RNA-silencing suppression by Tomato aspermy virus protein 2b.EMBO Rep. 2008 Aug;9(8):754-60. doi: 10.1038/embor.2008.118. Epub 2008 Jul 4. EMBO Rep. 2008. PMID: 18600235 Free PMC article.
-
A multifunctional human Argonaute2-specific monoclonal antibody.RNA. 2008 Jun;14(6):1244-53. doi: 10.1261/rna.973808. Epub 2008 Apr 22. RNA. 2008. PMID: 18430891 Free PMC article.
References
-
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337. - PubMed
-
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Ladgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MLL1 protein in mouse testis. Nature. 2006;442:203. - PubMed
-
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281. - PubMed
-
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356. - PubMed
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
