How does RNA editing affect dsRNA-mediated gene silencing?
- PMID: 17381308
- PMCID: PMC2846461
- DOI: 10.1101/sqb.2006.71.037
How does RNA editing affect dsRNA-mediated gene silencing?
Abstract
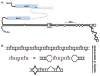
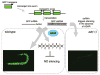
In general, double-stranded RNA (dsRNA)-binding proteins (dsRBPs) are not sequence-specific. A dsRNA molecule in a cell will interact with any dsRBP it comes in contact with, suggesting that different dsRNA-mediated pathways intersect and affect each other. This paper analyzes evidence that the ADAR RNA editing enzymes, which act on dsRNA, affect dsRNA-mediated gene silencing pathways. Examples of how ADARs alter gene silencing pathways such as RNA interference, as well as mechanisms that allow the pathways to coexist and maintain their unique functions, are discussed.
Figures
Similar articles
-
The role of RNA editing by ADARs in RNAi.Mol Cell. 2002 Oct;10(4):809-17. doi: 10.1016/s1097-2765(02)00649-4. Mol Cell. 2002. PMID: 12419225
-
ADBP-1 regulates an ADAR RNA-editing enzyme to antagonize RNA-interference-mediated gene silencing in Caenorhabditis elegans.Genetics. 2008 Oct;180(2):785-96. doi: 10.1534/genetics.108.093310. Epub 2008 Sep 9. Genetics. 2008. PMID: 18780728 Free PMC article.
-
Noncoding regions of C. elegans mRNA undergo selective adenosine to inosine deamination and contain a small number of editing sites per transcript.RNA Biol. 2015;12(2):162-74. doi: 10.1080/15476286.2015.1017220. RNA Biol. 2015. PMID: 25826568 Free PMC article.
-
Biology and Mechanisms of Short RNAs in Caenorhabditis elegans.Adv Genet. 2013;83:1-69. doi: 10.1016/B978-0-12-407675-4.00001-8. Adv Genet. 2013. PMID: 23890211 Review.
-
To protect and modify double-stranded RNA - the critical roles of ADARs in development, immunity and oncogenesis.Crit Rev Biochem Mol Biol. 2021 Feb;56(1):54-87. doi: 10.1080/10409238.2020.1856768. Epub 2020 Dec 27. Crit Rev Biochem Mol Biol. 2021. PMID: 33356612 Free PMC article. Review.
Cited by
-
Disruption in A-to-I Editing Levels Affects C. elegans Development More Than a Complete Lack of Editing.Cell Rep. 2019 Apr 23;27(4):1244-1253.e4. doi: 10.1016/j.celrep.2019.03.095. Cell Rep. 2019. PMID: 31018137 Free PMC article.
-
dsRNA expression in the mouse elicits RNAi in oocytes and low adenosine deamination in somatic cells.Nucleic Acids Res. 2012 Jan;40(1):399-413. doi: 10.1093/nar/gkr702. Epub 2011 Sep 8. Nucleic Acids Res. 2012. PMID: 21908396 Free PMC article.
-
Regulated RNA editing and functional epistasis in Shaker potassium channels.J Gen Physiol. 2009 Jan;133(1):17-27. doi: 10.1085/jgp.200810133. J Gen Physiol. 2009. PMID: 19114634 Free PMC article.
-
A-to-I RNA editing promotes developmental stage-specific gene and lncRNA expression.Genome Res. 2017 Mar;27(3):462-470. doi: 10.1101/gr.211169.116. Epub 2016 Dec 28. Genome Res. 2017. PMID: 28031250 Free PMC article.
-
Single-nucleotide polymorphisms among microRNA: big effects on cancer.Chin J Cancer. 2011 Jun;30(6):381-91. doi: 10.5732/cjc.011.10142. Chin J Cancer. 2011. PMID: 21627860 Free PMC article. Review.
References
-
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
