Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis
- PMID: 17384169
- PMCID: PMC1867373
- DOI: 10.1105/tpc.106.048009
Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis
Abstract
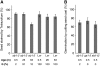
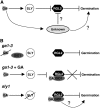
We explore the roles of gibberellin (GA) signaling genes SLEEPY1 (SLY1) and RGA-LIKE2 (RGL2) in regulation of seed germination in Arabidopsis thaliana, a plant in which the hormone GA is required for seed germination. Seed germination failure in the GA biosynthesis mutant ga1-3 is rescued by GA and by mutations in the DELLA gene RGL2, suggesting that RGL2 represses seed germination. RGL2 protein disappears before wild-type seed germination, consistent with the model that GA stimulates germination by causing the SCF(SLY1) E3 ubiquitin ligase complex to trigger ubiquitination and destruction of RGL2. Unlike ga1-3, the GA-insensitive sly1 mutants show variable seed dormancy. Seed lots with high seed dormancy after-ripened slowly, with stronger alleles requiring more time. We expected that if RGL2 negatively controls seed germination, sly1 mutant seeds that germinate well should accumulate lower RGL2 levels than those failing to germinate. Surprisingly, RGL2 accumulated at high levels even in after-ripened sly1 mutant seeds with 100% germination, suggesting that RGL2 disappearance is not a prerequisite for seed germination in the sly1 background. Without GA, several GA-induced genes show increased accumulation in sly1 seeds compared with ga1-3. It is possible that the RGL2 repressor of seed germination is inactivated by after-ripening of sly1 mutant seeds.
Figures







References
-
- An, Y.Q., McDowell, J.M., Huang, S., McKinney, E.C., Chambliss, S., and Meagher, R.B. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10 107–121. - PubMed
-
- Bewley, J.D., and Black, M. (1994). Seeds: Physiology of Development and Germination. (New York: Plenum Press).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

