Detection of mutant BRAF alleles in the plasma of patients with metastatic melanoma
- PMID: 17384209
- PMCID: PMC1867452
- DOI: 10.2353/jmoldx.2007.060135
Detection of mutant BRAF alleles in the plasma of patients with metastatic melanoma
Abstract
Mutations in the BRAF oncogene at amino acid 600 have been reported in 40 to 70% of human metastatic melanoma tissues, and the critical role of BRAF in the biology of melanoma has been established. Sampling the blood compartment to detect the mutational status of a solid tumor represents a highly innovative advance in cancer medicine, and such an approach could have advantages over tissue-based techniques. We report the development of a fluorescence-based polymerase chain reaction (PCR) assay to detect mutant BRAF alleles in plasma. A mutant-specific PCR assay was optimized to specifically amplify the mutant BRAF allele without amplifying the wild-type allele. Experiments mixing DNA from a BRAF mutant melanoma cell line with wild-type human placental DNA in varying proportions were performed to determine the threshold of this assay and to compare it with routine DNA sequencing. The assay was then applied to tissue and plasma specimens from patients with metastatic melanoma. The assay detected 0.1 ng of mutant DNA mixed in 100 ng of wild-type DNA and was 500-fold more sensitive than DNA sequencing. The assay detected mutant BRAF alleles in plasma samples from 14 of 26 (54%) metastatic melanoma patients. These data demonstrate the feasibility of blood-based testing for BRAF mutations in metastatic melanoma patients.
Figures

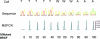
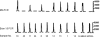
References
-
- Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, Moses TY, Hostetter G, Wagner U, Kakareka J, Salem G, Pohida T, Heenan P, Duray P, Kallioniemi O, Hayward NK, Trent JM, Meltzer PS. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. - PubMed
-
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. - PubMed
-
- Gorden A, Osman I, Gai W, He D, Huang W, Davidson A, Houghton AN, Busam K, Polsky D. Analysis of BRAF and N-RAS mutations in metastatic melanoma tissues. Cancer Res. 2003;63:3955–3957. - PubMed
-
- Kumar R, Angelini S, Czene K, Sauroja I, Hahka-Kemppinen M, Pyrhonen S, Hemminki K. BRAF mutations in metastatic melanoma: a possible association with clinical outcome. Clin Cancer Res. 2003;9:3362–3368. - PubMed
-
- Shinozaki M, Fujimoto A, Morton DL, Hoon DS. Incidence of BRAF oncogene mutation and clinical relevance for primary cutaneous melanomas. Clin Cancer Res. 2004;10:1753–1757. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

