Analysis of T-cell receptor-gamma gene rearrangements using oligonucleotide microchip: a novel approach for the determination of T-cell clonality
- PMID: 17384218
- PMCID: PMC1867449
- DOI: 10.2353/jmoldx.2007.060087
Analysis of T-cell receptor-gamma gene rearrangements using oligonucleotide microchip: a novel approach for the determination of T-cell clonality
Abstract
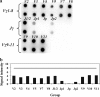
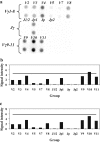
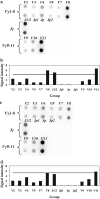
T-cell clonality estimation is important for the differential diagnosis between malignant and nonmalignant T-cell proliferation. Routinely used methods include polymerase chain reaction (PCR) analysis of T-cell receptor-gamma (TCR-gamma) gene rearrangements followed by Genescan analysis, polyacrylamide gel electrophoresis, or heteroduplex analysis to visualize amplification products. Here, we present a new method for the analysis after PCR of TCR-gamma rearrangements using hybridization on oligonucleotide microchip. A microchip was designed to contain specific probes for all functional variable (V) and joining (J) gene segments involved in rearrangements of the TCR-gamma locus. Fluorescently labeled fragments of rearranged gamma-chain from patients and donors were obtained in a multiplex nested PCR and hybridized with a microchip. The results were detected using a portable microchip analyzer. Samples from 49 patients with T-cell lymphomas or leukemias and 47 donors were analyzed for T-cell clonality by microchip and single-strand conformation polymorphism analysis, which served as a standard reference method. Comparison of two techniques showed full concordance of the results. The microchip-based approach also allowed the identification of V and J gene segments involved in the particular TCR-gamma rearrangement. The sensitivity of the method is sufficient to determine 10% of clonal cells in the sample.
Figures


Similar articles
-
Comparative analysis of TCR-gamma gene rearrangements by Genescan and polyacrylamide gel-electrophoresis in cutaneous T-cell lymphoma.Acta Derm Venereol. 2004;84(1):6-11. doi: 10.1080/00015550310015798. Acta Derm Venereol. 2004. PMID: 15040470
-
Pseudo-spikes are common in histologically benign lymphoid tissues.J Mol Diagn. 2000 Aug;2(3):145-52. doi: 10.1016/S1525-1578(10)60630-7. J Mol Diagn. 2000. PMID: 11229519 Free PMC article.
-
A novel clonality assay for the assessment of canine T cell proliferations.Vet Immunol Immunopathol. 2012 Jan 15;145(1-2):410-9. doi: 10.1016/j.vetimm.2011.12.019. Epub 2011 Dec 28. Vet Immunol Immunopathol. 2012. PMID: 22237398
-
[Analyses of the rearrangement of T-cell receptor- and immunoglobulin genes in the diagnosis of lymphoproliferative disorders].Veroff Pathol. 1995;144:1-109. Veroff Pathol. 1995. PMID: 7856305 Review. German.
-
Detection of clonal immunoglobulin and T-cell receptor gene recombination in hematological malignancies: monitoring minimal residual disease.Cardiovasc Hematol Disord Drug Targets. 2009 Jun;9(2):124-35. doi: 10.2174/187152909788488627. Cardiovasc Hematol Disord Drug Targets. 2009. PMID: 19519371 Review.
Cited by
-
Diagnostic microarrays in hematologic oncology: applications of high- and low-density arrays.Mol Diagn Ther. 2009;13(2):91-102. doi: 10.1007/BF03256318. Mol Diagn Ther. 2009. PMID: 19537844 Review.
-
Malleable immunoglobulin genes and hematopathology - the good, the bad, and the ugly: a paper from the 2007 William Beaumont hospital symposium on molecular pathology.J Mol Diagn. 2008 Sep;10(5):396-410. doi: 10.2353/jmoldx.2008.080061. Epub 2008 Aug 7. J Mol Diagn. 2008. PMID: 18687793 Free PMC article. Review.
-
Comparison of BIOMED-2 versus laboratory-developed polymerase chain reaction assays for detecting T-cell receptor-gamma gene rearrangements.J Mol Diagn. 2010 Mar;12(2):226-37. doi: 10.2353/jmoldx.2010.090042. J Mol Diagn. 2010. PMID: 20181819 Free PMC article.
References
-
- Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. Lyon: IARC Press; Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. World Health Organization Classification of Tumours. 2001
-
- Macintyre EA, Delabesse E. Molecular approaches to the diagnosis and evaluation of lymphoid malignancies. Semin Hematol. 1999;36:373–389. - PubMed
-
- Spagnolo DV, Ellis DW, Juneja S, Leong AS, Miliauskas J, Norris DL, Turner J. The role of molecular studies in lymphoma diagnosis: a review. Pathology. 2004;36:19–44. - PubMed
-
- Alexandre D, Lefranc MP. The human gamma/delta + and alpha/beta + T cells: a branched pathway of differentiation. Mol Immunol. 1992;29:447–451. - PubMed
-
- Ponti R, Quaglino P, Novelli M, Fierro MT, Comessatti A, Peroni A, Bonello L, Bernengo MG. T-cell receptor gamma gene rearrangement by multiplex polymerase chain reaction/heteroduplex analysis in patients with cutaneous T-cell lymphoma (mycosis fungoides/Sezary syndrome) and benign inflammatory disease: correlation with clinical, histological and immunophenotypical findings. Br J Dermatol. 2005;153:565–573. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

