Software note: using probe secondary structure information to enhance Affymetrix GeneChip background estimates
- PMID: 17387043
- PMCID: PMC2043163
- DOI: 10.1016/j.compbiolchem.2007.02.008
Software note: using probe secondary structure information to enhance Affymetrix GeneChip background estimates
Abstract
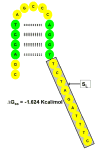
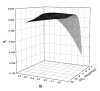
High-density short oligonucleotide microarrays are a primary research tool for assessing global gene expression. Background noise on microarrays comprises a significant portion of the measured raw data. A number of statistical techniques have been developed to correct for this background noise. Here, we demonstrate that probe minimum folding energy and structure can be used to enhance a previously existing model for background noise correction. We estimate that probe secondary structure accounts for up to 3% of all variation on Affymetrix microarrays.
Figures



Similar articles
-
Background correction using dinucleotide affinities improves the performance of GCRMA.BMC Bioinformatics. 2008 Oct 23;9:452. doi: 10.1186/1471-2105-9-452. BMC Bioinformatics. 2008. PMID: 18947404 Free PMC article.
-
Interactively optimizing signal-to-noise ratios in expression profiling: project-specific algorithm selection and detection p-value weighting in Affymetrix microarrays.Bioinformatics. 2004 Nov 1;20(16):2534-44. doi: 10.1093/bioinformatics/bth280. Epub 2004 Apr 29. Bioinformatics. 2004. PMID: 15117752
-
Expression profiling using affymetrix genechip probe arrays.Methods Mol Biol. 2007;366:13-40. doi: 10.1007/978-1-59745-030-0_2. Methods Mol Biol. 2007. PMID: 17568117
-
Expression Profiling Using Affymetrix GeneChip Microarrays.Methods Mol Biol. 2009;509:35-46. doi: 10.1007/978-1-59745-372-1_3. Methods Mol Biol. 2009. PMID: 19212713 Review.
-
Gene expression microarrays.Methods Mol Med. 2003;85:239-55. doi: 10.1385/1-59259-380-1:239. Methods Mol Med. 2003. PMID: 12710212 Review. No abstract available.
Cited by
-
Correction of spatial bias in oligonucleotide array data.Adv Bioinformatics. 2013;2013:167915. doi: 10.1155/2013/167915. Epub 2013 Mar 13. Adv Bioinformatics. 2013. PMID: 23573083 Free PMC article.
-
Regulatory divergence in Drosophila melanogaster and D. simulans, a genomewide analysis of allele-specific expression.Genetics. 2009 Oct;183(2):547-61, 1SI-21SI. doi: 10.1534/genetics.109.105957. Epub 2009 Aug 10. Genetics. 2009. PMID: 19667135 Free PMC article.
-
Design and analysis of mismatch probes for long oligonucleotide microarrays.BMC Genomics. 2008 Oct 17;9:491. doi: 10.1186/1471-2164-9-491. BMC Genomics. 2008. PMID: 18928550 Free PMC article.
-
Sequence characteristics define trade-offs between on-target and genome-wide off-target hybridization of oligoprobes.PLoS One. 2018 Jun 21;13(6):e0199162. doi: 10.1371/journal.pone.0199162. eCollection 2018. PLoS One. 2018. PMID: 29928000 Free PMC article.
-
ArrayInitiative - a tool that simplifies creating custom Affymetrix CDFs.BMC Bioinformatics. 2011 May 6;12:136. doi: 10.1186/1471-2105-12-136. BMC Bioinformatics. 2011. PMID: 21548938 Free PMC article.
References
-
- Anthony RM, Schuitema AR, Chan AB, Boender PJ, Klatser PR, Oskam L. Effect of secondary structure on single nucleotide polymorphism detection with a porous microarray matrix; implications for probe selection. Biotechniques. 2003;34:1082–1089. - PubMed
-
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

