Histone tails and the H3 alphaN helix regulate nucleosome mobility and stability
- PMID: 17387148
- PMCID: PMC1900026
- DOI: 10.1128/MCB.02229-06
Histone tails and the H3 alphaN helix regulate nucleosome mobility and stability
Abstract
Nucleosomes fulfill the apparently conflicting roles of compacting DNA within eukaryotic genomes while permitting access to regulatory factors. Central to this is their ability to stably associate with DNA while retaining the ability to undergo rearrangements that increase access to the underlying DNA. Here, we have studied different aspects of nucleosome dynamics including nucleosome sliding, histone dimer exchange, and DNA wrapping within nucleosomes. We find that alterations to histone proteins, especially the histone tails and vicinity of the histone H3 alphaN helix, can affect these processes differently, suggesting that they are mechanistically distinct. This raises the possibility that modifications to histone proteins may provide a means of fine-tuning specific aspects of the dynamic properties of nucleosomes to the context in which they are located.
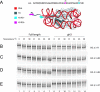
Figures






References
-
- Anderson, J. D., P. T. Lowary, and J. Widom. 2001. Effects of histone acetylation on the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 307:977-985. - PubMed
-
- Ausio, J., F. Dong, and K. E. van Holde. 1989. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone tails in the stabilization of the nucleosome. J. Mol. Biol. 206:451-463. - PubMed
-
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. - PubMed
-
- Bednar, J., R. A. Horowitz, S. A. Grigoryev, L. M. Carruthers, J. C. Hansen, A. J. Koster, and C. L. Woodcock. 1998. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. USA 95:14173-14178. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
