Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry
- PMID: 17389044
- PMCID: PMC1851972
- DOI: 10.1186/1471-2105-8-105
Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry
Abstract
Background: Structure elucidation of unknown small molecules by mass spectrometry is a challenge despite advances in instrumentation. The first crucial step is to obtain correct elemental compositions. In order to automatically constrain the thousands of possible candidate structures, rules need to be developed to select the most likely and chemically correct molecular formulas.
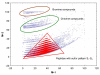
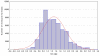
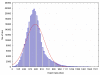
Results: An algorithm for filtering molecular formulas is derived from seven heuristic rules: (1) restrictions for the number of elements, (2) LEWIS and SENIOR chemical rules, (3) isotopic patterns, (4) hydrogen/carbon ratios, (5) element ratio of nitrogen, oxygen, phosphor, and sulphur versus carbon, (6) element ratio probabilities and (7) presence of trimethylsilylated compounds. Formulas are ranked according to their isotopic patterns and subsequently constrained by presence in public chemical databases. The seven rules were developed on 68,237 existing molecular formulas and were validated in four experiments. First, 432,968 formulas covering five million PubChem database entries were checked for consistency. Only 0.6% of these compounds did not pass all rules. Next, the rules were shown to effectively reducing the complement all eight billion theoretically possible C, H, N, S, O, P-formulas up to 2000 Da to only 623 million most probable elemental compositions. Thirdly 6,000 pharmaceutical, toxic and natural compounds were selected from DrugBank, TSCA and DNP databases. The correct formulas were retrieved as top hit at 80-99% probability when assuming data acquisition with complete resolution of unique compounds and 5% absolute isotope ratio deviation and 3 ppm mass accuracy. Last, some exemplary compounds were analyzed by Fourier transform ion cyclotron resonance mass spectrometry and by gas chromatography-time of flight mass spectrometry. In each case, the correct formula was ranked as top hit when combining the seven rules with database queries.
Conclusion: The seven rules enable an automatic exclusion of molecular formulas which are either wrong or which contain unlikely high or low number of elements. The correct molecular formula is assigned with a probability of 98% if the formula exists in a compound database. For truly novel compounds that are not present in databases, the correct formula is found in the first three hits with a probability of 65-81%. Corresponding software and supplemental data are available for downloads from the authors' website.
Figures




References
-
- Djerassi C, Silva CJ. Sponge Sterols - Origin and Biosynthesis. Accounts of Chemical Research. 1991;24:371–378.
-
- Omura S. Trends in the Search for Bioactive Microbial Metabolites. Journal of Industrial Microbiology. 1992;10:135–156. - PubMed
-
- Wray V. Carbon-Carbon Coupling-Constants - Compilation of Data and a Practical Guide. Progress in Nuclear Magnetic Resonance Spectroscopy. 1979;13:177–256.
-
- Buchanan BG, Smith DH, White WC, Gritter RJ, Feigenbaum EA, Lederberg J, Djerassi C. Applications of Artificial Intelligence for Chemical Inference .22. Automatic Rule Formation in Mass-Spectrometry by Means of Meta-Dendral Program. J Am Chem Soc. 1976;98:6168–6178.
-
- Olson DL, Norcross JA, O'Neil-Johnson M, Molitor PF, Detlefsen DJ, Wilson AG, Peck TL. Microflow NMR: concepts and capabilities. Anal Chem. 2004;76:2966–2974. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

