Recurrent DNA inversion rearrangements in the human genome
- PMID: 17389356
- PMCID: PMC1832218
- DOI: 10.1073/pnas.0701631104
Recurrent DNA inversion rearrangements in the human genome
Abstract
Several lines of evidence suggest that reiterated sequences in the human genome are targets for nonallelic homologous recombination (NAHR), which facilitates genomic rearrangements. We have used a PCR-based approach to identify breakpoint regions of rearranged structures in the human genome. In particular, we have identified intrachromosomal identical repeats that are located in reverse orientation, which may lead to chromosomal inversions. A bioinformatic workflow pathway to select appropriate regions for analysis was developed. Three such regions overlapping with known human genes, located on chromosomes 3, 15, and 19, were analyzed. The relative proportion of wild-type to rearranged structures was determined in DNA samples from blood obtained from different, unrelated individuals. The results obtained indicate that recurrent genomic rearrangements occur at relatively high frequency in somatic cells. Interestingly, the rearrangements studied were significantly more abundant in adults than in newborn individuals, suggesting that such DNA rearrangements might start to appear during embryogenesis or fetal life and continue to accumulate after birth. The relevance of our results in regard to human genomic variation is discussed.
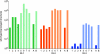
Figures







References
-
- Alt F, Kellems RE, Bertino JR, Schimke RT. J Biol Chem. 1978;253:1357–1370. - PubMed
-
- Schimke RT. Cell. 1984;37:705–713. - PubMed
-
- Shaw CJ, Lupski JR. Hum Mol Genet. 2004;13:R57–R64. - PubMed
-
- Higgs DR, Old JM, Pressley L, Clegg JB, Weatherall DJ. Nature. 1980;284:632–635. - PubMed
-
- Stankiewicz P, Lupski JR. Curr Opin Genet. 2002;12:312–319. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

