Structural insights into the degradation of Mcl-1 induced by BH3 domains
- PMID: 17389404
- PMCID: PMC1851040
- DOI: 10.1073/pnas.0701297104
Structural insights into the degradation of Mcl-1 induced by BH3 domains
Abstract
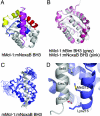
Apoptosis is held in check by prosurvival proteins of the Bcl-2 family. The distantly related BH3-only proteins bind to and antagonize them, thereby promoting apoptosis. Whereas binding of the BH3-only protein Noxa to prosurvival Mcl-1 induces Mcl-1 degradation by the proteasome, binding of another BH3-only ligand, Bim, elevates Mcl-1 protein levels. We compared the three-dimensional structures of the complexes formed between BH3 peptides of both Bim and Noxa, and we show that a discrete C-terminal sequence of the Noxa BH3 is necessary to instigate Mcl-1 degradation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Adams JM. Genes Dev. 2003;17:2481–2495. - PubMed
-
- Muchmore SW, Sattler M, Liang H, Meadows RP, Harlan JE, Yoon HS, Nettesheim D, Chang BS, Thompson CB, Wong S, et al. Nature. 1996;381:335–341. - PubMed
-
- Day CL, Chen L, Richardson SJ, Harrison PJ, Huang DCS, Hinds MG. J Biol Chem. 2005;280:4738–4744. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

