Ribosomal RNA guanine-(N2)-methyltransferases and their targets
- PMID: 17389639
- PMCID: PMC1874633
- DOI: 10.1093/nar/gkm104
Ribosomal RNA guanine-(N2)-methyltransferases and their targets
Abstract
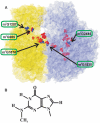
Five nearly universal methylated guanine-(N2) residues are present in bacterial rRNA in the ribosome. To date four out of five ribosomal RNA guanine-(N2)-methyltransferases are described. RsmC(YjjT) methylates G1207 of the 16S rRNA. RlmG(YgjO) and RlmL(YcbY) are responsible for the 23S rRNA m(2)G1835 and m(2)G2445 formation, correspondingly. RsmD(YhhF) is necessary for methylation of G966 residue of 16S rRNA. Structure of Escherichia coli RsmD(YhhF) methyltransferase and the structure of the Methanococcus jannaschii RsmC ortholog were determined. All ribosomal guanine-(N2)-methyltransferases have similar AdoMet-binding sites. In relation to the ribosomal substrate recognition, two enzymes that recognize assembled subunits are relatively small single domain proteins and two enzymes that recognize naked rRNA are larger proteins containing separate methyltransferase- and RNA-binding domains. The model for recognition of specific target nucleotide is proposed. The hypothetical role of the m(2)G residues in rRNA is discussed.
Figures
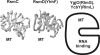
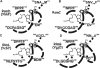


References
-
- Andersen NM, Douthwaite S. YebU is a m5C methyltransferase specific for 16S rRNA nucleotide 1407. J. Mol. Biol. 2006;359:777–786. - PubMed
-
- Mueller F, Brimacombe R. A new model for the three-dimensional folding of Escherichia coli 16S Ribosomal RNA. I. Fitting the RNA to a 3D electron microscopic map at 20 Å. J. Mol. Biol. 1997;271:524–544. - PubMed
-
- Cundliffe E. Ribosomal modification and resistance in antibiotic-producing organisms. Biochem. Soc. Symp. 1987;53:1–8. - PubMed
-
- Maravic G. Macrolide resistance based on the Erm-mediated rRNA methylation. Curr. Drug Targets Infect. Disord. 2004;4:193–202. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

