Real-time PCR mapping of DNaseI-hypersensitive sites using a novel ligation-mediated amplification technique
- PMID: 17389645
- PMCID: PMC1885650
- DOI: 10.1093/nar/gkm108
Real-time PCR mapping of DNaseI-hypersensitive sites using a novel ligation-mediated amplification technique
Abstract
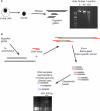
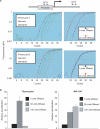
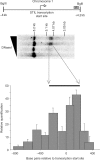
Mapping sites within the genome that are hypersensitive to digestion with DNaseI is an important method for identifying DNA elements that regulate transcription. The standard approach to locating these DNaseI-hypersensitive sites (DHSs) has been to use Southern blotting techniques, although we, and others, have recently published alternative methods using a range of technologies including high-throughput sequencing and genomic array tiling paths. In this article, we describe a novel protocol to use real-time PCR to map DHS. Advantages of the technique reported here include the small cell numbers required for each analysis, rapid, relatively low-cost experiments with minimal need for specialist equipment. Presented examples include comparative DHS mapping of known TAL1/SCL regulatory elements between human embryonic stem cells and K562 cells.
Figures

References
-
- Vyas P, Vickers MA, Simmons DL, Ayyub H, Craddock CF, Higgs DR. Cis-acting sequences regulating expression of the human alpha-globin cluster lie within constitutively open chromatin. Cell. 1992;69:781–793. - PubMed
-
- Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. - PubMed
-
- Ansel KM, Greenwald RJ, Agarwal S, Bassing CH, Monticelli S, Interlandi J, Djuretic IM, Lee DU, Sharpe AH, et al. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nat. Immunol. 2004;5:1251–1259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

