USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products
- PMID: 17389646
- PMCID: PMC1874642
- DOI: 10.1093/nar/gkm106
USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products
Abstract
We present a method that allows simultaneous fusion and cloning of multiple PCR products in a rapid and efficient manner. The procedure is based on the use of PCR primers that contain a single deoxyuridine residue near their 5' end. Treatment of the PCR products with a commercial deoxyuridine-excision reagent generates long 3' overhangs designed to specifically complement each other. The combination of this principle with the improved USER cloning technique provides a simple, fast and very efficient method to simultaneously fuse and clone multiple PCR fragments into a vector of interest. Around 90% positive clones were obtained when three different PCR products were fused and cloned into a USER-compatible vector in a simple procedure that, apart from the single PCR amplification step and the bacterial transformation, took approximately one hour. We expect this method to replace overlapping PCR and the use of type IIS restriction enzymes in many of their applications.
Figures
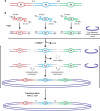


References
-
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. - PubMed
-
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

