Distance constraints between microRNA target sites dictate efficacy and cooperativity
- PMID: 17389647
- PMCID: PMC1874663
- DOI: 10.1093/nar/gkm133
Distance constraints between microRNA target sites dictate efficacy and cooperativity
Abstract
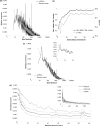
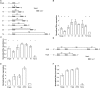
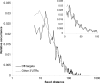
MicroRNAs (miRNAs) have the potential to regulate the expression of thousands of genes, but the mechanisms that determine whether a gene is targeted or not are poorly understood. We studied the genomic distribution of distances between pairs of identical miRNA seeds and found a propensity for moderate distances greater than about 13 nt between seed starts. Experimental data show that optimal down-regulation is obtained when two seed sites are separated by between 13 and 35 nt. By analyzing the distance between seed sites of endogenous miRNAs and transfected small interfering RNAs (siRNAs), we also find that cooperative targeting of sites with a separation in the optimal range can explain some of the siRNA off-target effects that have been reported in the literature.
Figures
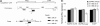
References
-
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. - PubMed
-
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. - PubMed
-
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. - PubMed
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
-
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

