Accumulation of mitochondrial DNA deletion mutations in aged muscle fibers: evidence for a causal role in muscle fiber loss
- PMID: 17389720
- PMCID: PMC2846622
- DOI: 10.1093/gerona/62.3.235
Accumulation of mitochondrial DNA deletion mutations in aged muscle fibers: evidence for a causal role in muscle fiber loss
Abstract
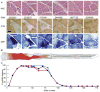
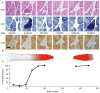
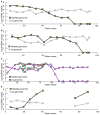
Although mitochondrial mutation abundance has been recognized to increase in an age-dependent manner, the impact of mutation has been more difficult to establish. Using quantitative polymerase chain reaction, we measured the intracellular abundance of mutant and wild-type mitochondrial genomes along the length of individual laser-captured microdissected muscle fibers from aged rat quadriceps. Aged muscle fibers possessed segmental, clonal intracellular expansions of unique somatically derived mitochondrial DNA (mtDNA) deletion mutations. When the mutation abundance surpassed 90% of the total mitochondrial genomes, the fiber lost cytochrome c oxidase activity and exhibited an increase in succinate dehydrogenase activity. In addition to the mitochondrial enzymatic abnormalities, some fibers displayed abnormal morphology such as fiber splitting, atrophy, and breakage. Deletion mutation accumulation was linked to these aberrant morphologies with more severe cellular pathologies resulting from higher deletion mutation abundance. In summary, our measurements indicate that age-induced mtDNA deletion mutations expand within individual muscle fibers, eliciting fiber dysfunction and breakage.
Figures

Similar articles
-
Mitochondrial DNA deletion mutations are concomitant with ragged red regions of individual, aged muscle fibers: analysis by laser-capture microdissection.Nucleic Acids Res. 2001 Nov 1;29(21):4502-8. doi: 10.1093/nar/29.21.4502. Nucleic Acids Res. 2001. PMID: 11691938 Free PMC article.
-
Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting, and oxidative damage in sarcopenia.FASEB J. 2001 Feb;15(2):322-32. doi: 10.1096/fj.00-0320com. FASEB J. 2001. PMID: 11156948
-
Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers.Am J Hum Genet. 2006 Sep;79(3):469-80. doi: 10.1086/507132. Epub 2006 Jul 7. Am J Hum Genet. 2006. PMID: 16909385 Free PMC article.
-
Mitochondrial DNA deletion mutations and sarcopenia.Ann N Y Acad Sci. 2002 Apr;959:412-23. doi: 10.1111/j.1749-6632.2002.tb02111.x. Ann N Y Acad Sci. 2002. PMID: 11976214 Review.
-
Influences of caloric restriction on age-associated skeletal muscle fiber characteristics and mitochondrial changes in rats and mice.Ann N Y Acad Sci. 1998 Nov 20;854:182-91. doi: 10.1111/j.1749-6632.1998.tb09901.x. Ann N Y Acad Sci. 1998. PMID: 9928429 Review.
Cited by
-
Analyses of Mitochondrial DNA and Immune Phenotyping Suggest Accelerated T-Cell Turnover in Treated HIV.J Acquir Immune Defic Syndr. 2018 Nov 1;79(3):399-406. doi: 10.1097/QAI.0000000000001824. J Acquir Immune Defic Syndr. 2018. PMID: 30312276 Free PMC article.
-
Age-dependent changes in mitochondrial morphology and volume are not predictors of lifespan.Aging (Albany NY). 2014 Feb;6(2):118-30. doi: 10.18632/aging.100639. Aging (Albany NY). 2014. PMID: 24642473 Free PMC article.
-
Quality matters: how does mitochondrial network dynamics and quality control impact on mtDNA integrity?Philos Trans R Soc Lond B Biol Sci. 2014 Jul 5;369(1646):20130442. doi: 10.1098/rstb.2013.0442. Philos Trans R Soc Lond B Biol Sci. 2014. PMID: 24864312 Free PMC article. Review.
-
Control of DNA integrity in skeletal muscle under physiological and pathological conditions.Cell Mol Life Sci. 2017 Oct;74(19):3439-3449. doi: 10.1007/s00018-017-2530-0. Epub 2017 Apr 25. Cell Mol Life Sci. 2017. PMID: 28444416 Free PMC article. Review.
-
The road to rack and ruin: selecting deleterious mitochondrial DNA variants.Philos Trans R Soc Lond B Biol Sci. 2014 Jul 5;369(1646):20130451. doi: 10.1098/rstb.2013.0451. Philos Trans R Soc Lond B Biol Sci. 2014. PMID: 24864317 Free PMC article. Review.
References
-
- Harman D. The biologic clock: the mitochondria? Journal of the American Geriatrics Society. 1972;20(4):145–7. - PubMed
-
- Miquel J, et al. Mitochondrial role in cell aging. Exp Geront. 1980;15:575–591. - PubMed
-
- Wallace DC. Mitochondrial Diseases in Man and Mouse. Science. 1999;283(5407):1482–1488. - PubMed
-
- Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–23. - PubMed
-
- Kujoth GC, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309(5733):481–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

