Large-scale comparative genomic ranking of taxonomically restricted genes (TRGs) in bacterial and archaeal genomes
- PMID: 17389915
- PMCID: PMC1824705
- DOI: 10.1371/journal.pone.0000324
Large-scale comparative genomic ranking of taxonomically restricted genes (TRGs) in bacterial and archaeal genomes
Abstract
Background: Lineage-specific, or taxonomically restricted genes (TRGs), especially those that are species and strain-specific, are of special interest because they are expected to play a role in defining exclusive ecological adaptations to particular niches. Despite this, they are relatively poorly studied and little understood, in large part because many are still orphans or only have homologues in very closely related isolates. This lack of homology confounds attempts to establish the likelihood that a hypothetical gene is expressed and, if so, to determine the putative function of the protein.
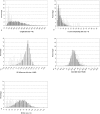
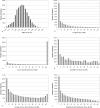
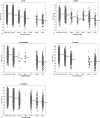
Methodology/principal findings: We have developed "QIPP" ("Quality Index for Predicted Proteins"), an index that scores the "quality" of a protein based on non-homology-based criteria. QIPP can be used to assign a value between zero and one to any protein based on comparing its features to other proteins in a given genome. We have used QIPP to rank the predicted proteins in the proteomes of Bacteria and Archaea. This ranking reveals that there is a large amount of variation in QIPP scores, and identifies many high-scoring orphans as potentially "authentic" (expressed) orphans. There are significant differences in the distributions of QIPP scores between orphan and non-orphan genes for many genomes and a trend for less well-conserved genes to have lower QIPP scores.
Conclusions: The implication of this work is that QIPP scores can be used to further annotate predicted proteins with information that is independent of homology. Such information can be used to prioritize candidates for further analysis. Data generated for this study can be found in the OrphanMine at http://www.genomics.ceh.ac.uk/orphan_mine.
Conflict of interest statement
Figures

References
-
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. - PubMed
-
- Wilson GA, Bertrand N, Patel Y, Hughes JB, Feil EJ, et al. Orphans as taxonomically restricted and ecologically important genes. Microbiology. 2005;151:2499–2501. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

