Mutational analysis of aspartoacylase: implications for Canavan disease
- PMID: 17391648
- PMCID: PMC1933483
- DOI: 10.1016/j.brainres.2007.02.069
Mutational analysis of aspartoacylase: implications for Canavan disease
Abstract
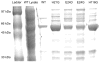
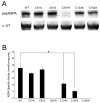
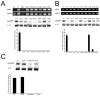
Mutations that result in near undetectable activity of aspartoacylase, which catalyzes the deacetylation of N-acetyl-l-aspartate, correlate with Canavan Disease, a neurodegenerative disorder usually fatal during childhood. The underlying biochemical mechanisms of how these mutations ablate activity are poorly understood. Therefore, we developed and tested a three-dimensional homology model of aspartoacylase based on zinc dependent carboxypeptidase A. Mutations of the putative zinc-binding residues (H21G, E24D/G, and H116G), the general proton donor (E178A), and mutants designed to switch the order of the zinc-binding residues (H21E/E24H and E24H/H116E) yielded wild-type aspartoacylase protein levels and undetectable ASPA activity. Mutations that affect substrate carboxyl binding (R71N) and transition state stabilization (R63N) also yielded wild-type aspartoacylase protein levels and undetectable aspartoacylase activity. Alanine substitutions of Cys124 and Cys152, residues indicated by homology modeling to be in close proximity and in the proper orientation for disulfide bonding, yielded reduced ASPA protein and activity levels. Finally, expression of several previously tested (E24G, D68A, C152W, E214X, D249V, E285A, and A305E) and untested (H21P, A57T, I143T, P183H, M195R, K213E/G274R, G274R, and F295S) Canavan Disease mutations resulted in undetectable enzyme activity, and only E285A and P183H showed wild-type aspartoacylase protein levels. These results show that aspartoacylase is a member of the caboxypeptidase A family and offer novel explanations for most loss-of-function aspartoacylase mutations associated with Canavan Disease.
Figures


References
-
- Auld DS. Zinc coordination sphere in biochemical zinc sites. Biometals. 2001;14:271–313. - PubMed
-
- Benuck M, D’Adamo AF., Jr Acetyl transport mechanisms. Metabolism of N-acetyl-L-aspartic acid in the non-nervous tissues of the rat. Biochim Biophys Acta. 1968;152:611–618. - PubMed
-
- Bukrinsky JT, Bjerrum MJ, Kadziola A. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. Biochemistry. 1998;37:16555–16564. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

