Structural basis for the recognition of blood group trisaccharides by norovirus
- PMID: 17392366
- PMCID: PMC1900264
- DOI: 10.1128/JVI.00219-07
Structural basis for the recognition of blood group trisaccharides by norovirus
Abstract
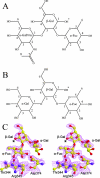
Noroviruses are one of the major causes of nonbacterial gastroenteritis epidemics in humans. Recent studies on norovirus receptors show that different noroviruses recognize different human histo-blood group antigens (HBGAs), and eight receptor binding patterns of noroviruses have been identified. The P domain of the norovirus capsids is directly involved in this recognition. To determine the precise locations and receptor binding modes of HBGA carbohydrates on the viral capsids, a recombinant P protein of a GII-4 strain norovirus, VA387, was cocrystallized with synthetic type A or B trisaccharides. Based on complex crystal structures observed at a 2.0-A resolution, we demonstrated that the receptor binding site lies at the outermost end of the P domain and forms an extensive hydrogen-bonding network with the saccharide ligand. The A and B trisaccharides display similar binding modes, and the common fucose ring plays a key role in this interaction. The extensive interface between the two protomers in a P dimer also plays a crucial role in the formation of the receptor binding interface.
Figures




References
-
- Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. - PubMed
-
- Huang, P., T. Farkas, S. Marionneau, W. Zhong, N. Ruvoen-Clouet, A. L. Morrow, M. Altaye, L. K. Pickering, D. S. Newburg, J. LePendu, and X. Jiang. 2003. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J. Infect. Dis. 188:19-31. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

