Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis
- PMID: 17394648
- PMCID: PMC1852104
- DOI: 10.1186/1471-2164-8-86
Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis
Abstract
Background: The Archaea are highly diverse in terms of their physiology, metabolism and ecology. Presently, very few molecular characteristics are known that are uniquely shared by either all archaea or the different main groups within archaea. The evolutionary relationships among different groups within the Euryarchaeota branch are also not clearly understood.
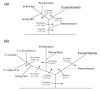
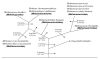
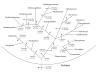
Results: We have carried out comprehensive analyses on each open reading frame (ORFs) in the genomes of 11 archaea (3 Crenarchaeota--Aeropyrum pernix, Pyrobaculum aerophilum and Sulfolobus acidocaldarius; 8 Euryarchaeota--Pyrococcus abyssi, Methanococcus maripaludis, Methanopyrus kandleri, Methanococcoides burtonii, Halobacterium sp. NCR-1, Haloquadratum walsbyi, Thermoplasma acidophilum and Picrophilus torridus) to search for proteins that are unique to either all Archaea or for its main subgroups. These studies have identified 1448 proteins or ORFs that are distinctive characteristics of Archaea and its various subgroups and whose homologues are not found in other organisms. Six of these proteins are unique to all Archaea, 10 others are only missing in Nanoarchaeum equitans and a large number of other proteins are specific for various main groups within the Archaea (e.g. Crenarchaeota, Euryarchaeota, Sulfolobales and Desulfurococcales, Halobacteriales, Thermococci, Thermoplasmata, all methanogenic archaea or particular groups of methanogens). Of particular importance is the observation that 31 proteins are uniquely present in virtually all methanogens (including M. kandleri) and 10 additional proteins are only found in different methanogens as well as A. fulgidus. In contrast, no protein was exclusively shared by various methanogen and any of the Halobacteriales or Thermoplasmatales. These results strongly indicate that all methanogenic archaea form a monophyletic group exclusive of other archaea and that this lineage likely evolved from Archaeoglobus. In addition, 15 proteins that are uniquely shared by M. kandleri and Methanobacteriales suggest a close evolutionary relationship between them. In contrast to the phylogenomics studies, a monophyletic grouping of archaea is not supported by phylogenetic analyses based on protein sequences.
Conclusion: The identified archaea-specific proteins provide novel molecular markers or signature proteins that are distinctive characteristics of Archaea and all of its major subgroups. The species distributions of these proteins provide novel insights into the evolutionary relationships among different groups within Archaea, particularly regarding the origin of methanogenesis. Most of these proteins are of unknown function and further studies should lead to discovery of novel biochemical and physiological characteristics that are unique to either all archaea or its different subgroups.
Figures

References
-
- Ludwig W, Klenk HP. Overview:A phylogenetic backbone and taxonomic framework for prokaryotic systamatics. In: D.R. B and R.W. C, editor. Bergey's Manual of Systematic Bacteriology. Berlin, Springer-Verlag; 2001. pp. 49–65.
-
- Skophammer RG, Herbold CW, Rivera MC, Servin JA, Lake JA. Evidence that the Root of the Tree of Life is not within the Archaea. Mol Biol Evol. 2006. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

