Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA
- PMID: 17396152
- PMCID: PMC1852787
- DOI: 10.1038/sj.emboj.7601661
Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA
Abstract
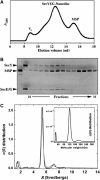
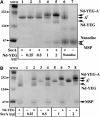
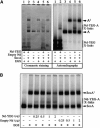
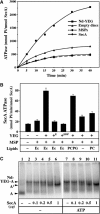
The translocon is a membrane-embedded protein assembly that catalyzes protein movement across membranes. The core translocon, the SecYEG complex, forms oligomers, but the protein-conducting channel is at the center of the monomer. Defining the properties of the SecYEG protomer is thus crucial to understand the underlying function of oligomerization. We report here the reconstitution of a single SecYEG complex into nano-scale lipid bilayers, termed Nanodiscs. These water-soluble particles allow one to probe the interactions of the SecYEG complex with its cytosolic partner, the SecA dimer, in a membrane-like environment. The results show that the SecYEG complex triggers dissociation of the SecA dimer, associates only with the SecA monomer and suffices to (pre)-activate the SecA ATPase. Acidic lipids surrounding the SecYEG complex also contribute to the binding affinity and activation of SecA, whereas mutations in the largest cytosolic loop of the SecY subunit, known to abolish the translocation reaction, disrupt both the binding and activation of SecA. Altogether, the results define the fundamental contribution of the SecYEG protomer in the translocation subreactions and illustrate the power of nanoscale lipid bilayers in analyzing the dynamics occurring at the membrane.
Figures


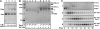

References
-
- Bassilana M, Arkowitz RA, Wickner W (1992) The role of the mature domain of proOmpA in the translocation ATPase reaction. J Biol Chem 267: 25246–25250 - PubMed
-
- Bayburt TH, Grinkova YV, Sligar SG (2006) Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch Biochem Biophys 450: 215–222 - PubMed
-
- Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G (2001) Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107: 361–372 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

