Analysis of meiotic recombination in 22q11.2, a region that frequently undergoes deletions and duplications
- PMID: 17397557
- PMCID: PMC1855045
- DOI: 10.1186/1471-2350-8-14
Analysis of meiotic recombination in 22q11.2, a region that frequently undergoes deletions and duplications
Abstract
Background: The 22q11.2 deletion syndrome is the most frequent genomic disorder with an estimated frequency of 1/4000 live births. The majority of patients (90%) have the same deletion of 3 Mb (Typically Deleted Region, TDR) that results from aberrant recombination at meiosis between region specific low-copy repeats (LCRs).
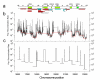
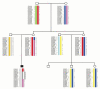
Methods: As a first step towards the characterization of recombination rates and breakpoints within the 22q11.2 region we have constructed a high resolution recombination breakpoint map based on pedigree analysis and a population-based historical recombination map based on LD analysis.
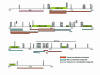
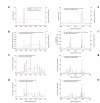
Results: Our pedigree map allows the location of recombination breakpoints with a high resolution (potential recombination hotspots), and this approach has led to the identification of 5 breakpoint segments of 50 kb or less (8.6 kb the smallest), that coincide with historical hotspots. It has been suggested that aberrant recombination leading to deletion (and duplication) is caused by low rates of Allelic Homologous Recombination (AHR) within the affected region. However, recombination rate estimates for 22q11.2 region show that neither average recombination rates in the 22q11.2 region or within LCR22-2 (the LCR implicated in most deletions and duplications), are significantly below chromosome 22 averages. Furthermore, LCR22-2, the repeat most frequently implicated in rearrangements, is also the LCR22 with the highest levels of AHR. In addition, we find recombination events in the 22q11.2 region to cluster within families. Within this context, the same chromosome recombines twice in one family; first by AHR and in the next generation by NAHR resulting in an individual affected with the del22q11.2 syndrome.
Conclusion: We show in the context of a first high resolution pedigree map of the 22q11.2 region that NAHR within LCR22 leading to duplications and deletions cannot be explained exclusively under a hypothesis of low AHR rates. In addition, we find that AHR recombination events cluster within families. If normal and aberrant recombination are mechanistically related, the fact that LCR22s undergo frequent AHR and that we find familial differences in recombination rates within the 22q11.2 region would have obvious health-related implications.
Figures


References
-
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, Ainscough R, Almeida JP, Babbage A, Bagguley C, Bailey J, Barlow K, Bates KN, Beasley O, Bird CP, Blakey S, Bridgeman AM, Buck D, Burgess J, Burrill WD, O'Brien KP, . The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. - DOI - PubMed
-
- Shaikh TH, Kurahashi H, Emanuel BS. Evolutionarily conserved low copy repeats (LCRs) in 22q11 mediate deletions, duplications, translocations, and genomic instability: an update and literature review. Genet Med. 2001;3:6–13. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

