Metabolic flux analysis in a nonstationary system: fed-batch fermentation of a high yielding strain of E. coli producing 1,3-propanediol
- PMID: 17400499
- PMCID: PMC2048574
- DOI: 10.1016/j.ymben.2007.01.003
Metabolic flux analysis in a nonstationary system: fed-batch fermentation of a high yielding strain of E. coli producing 1,3-propanediol
Abstract
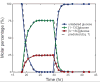
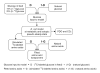
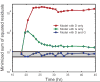
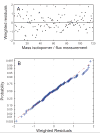
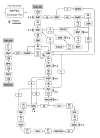
Metabolic fluxes estimated from stable-isotope studies provide a key to understanding cell physiology and regulation of metabolism. A limitation of the classical method for metabolic flux analysis (MFA) is the requirement for isotopic steady state. To extend the scope of flux determination from stationary to nonstationary systems, we present a novel modeling strategy that combines key ideas from isotopomer spectral analysis (ISA) and stationary MFA. Isotopic transients of the precursor pool and the sampled products are described by two parameters, D and G parameters, respectively, which are incorporated into the flux model. The G value is the fraction of labeled product in the sample, and the D value is the fractional contribution of the feed for the production of labeled products. We illustrate the novel modeling strategy with a nonstationary system that closely resembles industrial production conditions, i.e. fed-batch fermentation of Escherichia coli that produces 1,3-propanediol (PDO). Metabolic fluxes and the D and G parameters were estimated by fitting labeling distributions of biomass amino acids measured by GC/MS to a model of E. coli metabolism. We obtained highly consistent fits from the data with 82 redundant measurements. Metabolic fluxes were estimated for 20 time points during course of the fermentation. As such we established, for the first time, detailed time profiles of in vivo fluxes. We found that intracellular fluxes changed significantly during the fed-batch. The intracellular flux associated with PDO pathway increased by 10%. Concurrently, we observed a decrease in the split ratio between glycolysis and pentose phosphate pathway from 70/30 to 50/50 as a function of time. The TCA cycle flux, on the other hand, remained constant throughout the fermentation. Furthermore, our flux results provided additional insight in support of the assumed genotype of the organism.
Figures

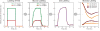


References
-
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. 2006a submitted to Anal Chem. - PubMed
-
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006b;8:324–337. - PubMed
-
- Brunengraber H, Kelleher JK, Des RC. Applications of mass isotopomer analysis to nutrition research. Annu Rev Nutr. 1997;17:559–596. - PubMed
-
- Dauner M, Sauer U. GC-MS analysis of amino acids rapidly provides rich information for isotopomer balancing. Biotechnol Prog. 2000;16:642–649. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

