Tracking single Kinesin molecules in the cytoplasm of mammalian cells
- PMID: 17400704
- PMCID: PMC1877757
- DOI: 10.1529/biophysj.106.100206
Tracking single Kinesin molecules in the cytoplasm of mammalian cells
Abstract
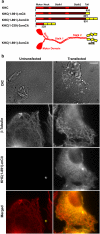
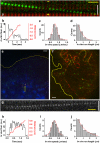
Understanding dynamic cellular processes requires precise knowledge of the distribution, transport, and interactions of individual molecules in living cells. Despite recent progress in in vivo imaging, it has not been possible to express and directly track single molecules in the cytoplasm of live cells. Here, we overcome these limitations by combining fluorescent protein-labeling with high resolution total internal reflection fluorescence microcopy, using the molecular motor Kinesin-1 as model system. First, we engineered a three-tandem monomeric Citrine tag for genetic labeling of individual molecules and expressed this motor in COS cells. Detailed analysis of the quantized photobleaching behavior of individual fluorescent spots demonstrates that we are indeed detecting single proteins in the cytoplasm of live cells. Tracking the movement of individual cytoplasmic molecules reveals that individual Kinesin-1 motors in vivo move with an average speed of 0.78 +/- 0.11 microm/s and display an average run length of 1.17 +/- 0.38 microm, which agrees well with in vitro measurements. Thus, Kinesin-1's speed and processivity are not upregulated or hindered by macromolecular crowding. Second, we demonstrate that standard deviation maps of the fluorescence intensity computed from single molecule image sequences can be used to reveal important physiological information about infrequent cellular events in the noisy fluorescence background of live cells. Finally, we show that tandem fluorescent protein tags enable single-molecule, in vitro analyses of extracted, mammalian-expressed proteins. Thus, by combining direct genetic labeling and single molecule imaging in vivo, our work establishes an important new biophysical method for observing single molecules expressed and localized in the mammalian cytoplasm.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

