Faithful modeling of transient expression and its application to elucidating negative feedback regulation
- PMID: 17400752
- PMCID: PMC1851052
- DOI: 10.1073/pnas.0611168104
Faithful modeling of transient expression and its application to elucidating negative feedback regulation
Abstract
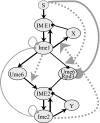
Modeling and analysis of genetic regulatory networks is essential both for better understanding their dynamic behavior and for elucidating and refining open issues. We hereby present a discrete computational model that effectively describes the transient and sequential expression of a network of genes in a representative developmental pathway. Our model system is a transcriptional cascade that includes positive and negative feedback loops directing the initiation and progression through meiosis in budding yeast. The computational model allows qualitative analysis of the transcription of early meiosis-specific genes, specifically, Ime2 and their master activator, Ime1. The simulations demonstrate a robust transcriptional behavior with respect to the initial levels of Ime1 and Ime2. The computational results were verified experimentally by deleting various genes and by changing initial conditions. The model has a strong predictive aspect, and it provides insights into how to distinguish among and reason about alternative hypotheses concerning the mode by which negative regulation through Ime1 and Ime2 is accomplished. Some predictions were validated experimentally, for instance, showing that the decline in the transcription of IME1 depends on Rpd3, which is recruited by Ime1 to its promoter. Finally, this general model promotes the analysis of systems that are devoid of consistent quantitative data, as is often the case, and it can be easily adapted to other developmental pathways.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
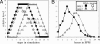


Similar articles
-
Glucose and nitrogen regulate the switch from histone deacetylation to acetylation for expression of early meiosis-specific genes in budding yeast.Mol Cell Biol. 2004 Jun;24(12):5197-208. doi: 10.1128/MCB.24.12.5197-5208.2004. Mol Cell Biol. 2004. PMID: 15169885 Free PMC article.
-
IME1 gene encodes a transcription factor which is required to induce meiosis in Saccharomyces cerevisiae.Dev Genet. 1994;15(2):139-47. doi: 10.1002/dvg.1020150204. Dev Genet. 1994. PMID: 8205723
-
GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast.Proc Natl Acad Sci U S A. 1999 Jun 8;96(12):6835-40. doi: 10.1073/pnas.96.12.6835. Proc Natl Acad Sci U S A. 1999. PMID: 10359799 Free PMC article.
-
Transcriptional regulation of meiosis in budding yeast.Int Rev Cytol. 2003;224:111-71. doi: 10.1016/s0074-7696(05)24004-4. Int Rev Cytol. 2003. PMID: 12722950 Review.
-
The Ime2 protein kinase family in fungi: more duties than just meiosis.Mol Microbiol. 2011 Apr;80(1):1-13. doi: 10.1111/j.1365-2958.2011.07575.x. Epub 2011 Mar 1. Mol Microbiol. 2011. PMID: 21306447 Review.
Cited by
-
The effective application of a discrete transition model to explore cell-cycle regulation in yeast.BMC Res Notes. 2013 Aug 6;6:311. doi: 10.1186/1756-0500-6-311. BMC Res Notes. 2013. PMID: 23915717 Free PMC article.
-
A switch from a gradient to a threshold mode in the regulation of a transcriptional cascade promotes robust execution of meiosis in budding yeast.PLoS One. 2010 Jun 8;5(6):e11005. doi: 10.1371/journal.pone.0011005. PLoS One. 2010. PMID: 20543984 Free PMC article.
-
Functional dissection of IME1 transcription using quantitative promoter-reporter screening.Genetics. 2010 Nov;186(3):829-41. doi: 10.1534/genetics.110.122200. Epub 2010 Aug 25. Genetics. 2010. PMID: 20739709 Free PMC article.
-
Interactions between the circadian clock and TGF-β signaling pathway in zebrafish.PLoS One. 2018 Jun 25;13(6):e0199777. doi: 10.1371/journal.pone.0199777. eCollection 2018. PLoS One. 2018. PMID: 29940038 Free PMC article.
-
Ime1 and Ime2 are required for pseudohyphal growth of Saccharomyces cerevisiae on nonfermentable carbon sources.Mol Cell Biol. 2010 Dec;30(23):5514-30. doi: 10.1128/MCB.00390-10. Epub 2010 Sep 27. Mol Cell Biol. 2010. PMID: 20876298 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

