Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting
- PMID: 17400893
- PMCID: PMC1867363
- DOI: 10.1105/tpc.107.050062
Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting
Abstract
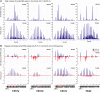
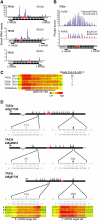
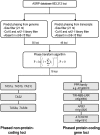
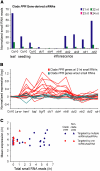
Posttranscriptional RNA silencing of many endogenous transcripts, viruses, and transgenes involves the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 (RDR6/DCL4)-dependent short interfering RNA (siRNA) biogenesis pathway. Arabidopsis thaliana contains several families of trans-acting siRNAs (tasiRNAs) that form in 21-nucleotide phased arrays through the RDR6/DCL4-dependent pathway and that negatively regulate target transcripts. Using deep sequencing technology and computational approaches, the phasing patterns of known tasiRNAs and tasiRNA-like loci from across the Arabidopsis genome were analyzed in wild-type plants and silencing-defective mutants. Several gene transcripts were found to be routed through the RDR6/DCL4-dependent pathway after initial targeting by one or multiple miRNAs or tasiRNAs, the most conspicuous example of which was an expanding clade of genes encoding pentatricopeptide repeat (PPR) proteins. Interestingly, phylogenetic analysis using Populus trichocarpa revealed evidence for small RNA-mediated regulatory mechanisms within a similarly expanded group of PPR genes. We suggest that posttranscriptional silencing mechanisms operate on an evolutionary scale to buffer the effects of rapidly expanding gene families.
Figures

Similar articles
-
Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins.RNA. 2007 Aug;13(8):1268-78. doi: 10.1261/rna.541307. Epub 2007 Jun 25. RNA. 2007. PMID: 17592042 Free PMC article.
-
The CaMV transactivator/viroplasmin interferes with RDR6-dependent trans-acting and secondary siRNA pathways in Arabidopsis.Nucleic Acids Res. 2008 Oct;36(18):5896-909. doi: 10.1093/nar/gkn590. Epub 2008 Sep 18. Nucleic Acids Res. 2008. PMID: 18801846 Free PMC article.
-
Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation.Cell. 2008 Apr 4;133(1):128-41. doi: 10.1016/j.cell.2008.02.033. Epub 2008 Mar 13. Cell. 2008. PMID: 18342362
-
miRNAs in the biogenesis of trans-acting siRNAs in higher plants.Semin Cell Dev Biol. 2010 Oct;21(8):798-804. doi: 10.1016/j.semcdb.2010.03.008. Epub 2010 Mar 30. Semin Cell Dev Biol. 2010. PMID: 20359543 Review.
-
[Trans-acting short interfering RNAs].Postepy Biochem. 2006;52(3):253-9. Postepy Biochem. 2006. PMID: 17201060 Review. Polish.
Cited by
-
High-throughput sequencing of RNA silencing-associated small RNAs in olive (Olea europaea L.).PLoS One. 2011;6(11):e27916. doi: 10.1371/journal.pone.0027916. Epub 2011 Nov 28. PLoS One. 2011. PMID: 22140484 Free PMC article.
-
The Influence of Genotype and Environment on Small RNA Profiles in Grapevine Berry.Front Plant Sci. 2016 Oct 5;7:1459. doi: 10.3389/fpls.2016.01459. eCollection 2016. Front Plant Sci. 2016. PMID: 27761135 Free PMC article.
-
Non-coding RNAs in the plant response to abiotic stress.Planta. 2012 Oct;236(4):943-58. doi: 10.1007/s00425-012-1693-z. Epub 2012 Jul 4. Planta. 2012. PMID: 22761008 Review.
-
Different gene families in Arabidopsis thaliana transposed in different epochs and at different frequencies throughout the rosids.Plant Cell. 2011 Dec;23(12):4241-53. doi: 10.1105/tpc.111.093567. Epub 2011 Dec 16. Plant Cell. 2011. PMID: 22180627 Free PMC article.
-
PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1.PLoS Genet. 2012;8(4):e1002616. doi: 10.1371/journal.pgen.1002616. Epub 2012 Apr 19. PLoS Genet. 2012. PMID: 22536158 Free PMC article.
References
-
- Adenot, X., Elmayan, T., Lauressergues, D., Boutet, S., Bouche, N., Gasciolli, V., and Vaucheret, H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16 927–932. - PubMed
-
- Akagi, H., Nakamura, A., Yokozeki-Misono, Y., Inagaki, A., Takahashi, H., Mori, K., and Fujimura, T. (2004). Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor. Appl. Genet. 108 1449–1457. - PubMed
-
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. - PubMed
-
- Allen, E., Xie, Z., Gustafson, A.M., Sung, G.H., Spatafora, J.W., and Carrington, J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36 1282–1290. - PubMed
-
- Aravin, A., et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442 203–207. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

