Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility
- PMID: 17400897
- PMCID: PMC1867369
- DOI: 10.1105/tpc.106.044123
Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility
Abstract
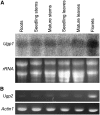
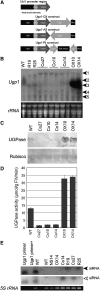
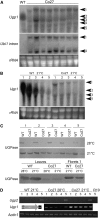
UDP-glucose pyrophosphorylase (UGPase) catalyzes the reversible production of glucose-1-phosphate and UTP to UDP-glucose and pyrophosphate. The rice (Oryza sativa) genome contains two homologous UGPase genes, Ugp1 and Ugp2. We report a functional characterization of rice Ugp1, which is expressed throughout the plant, with highest expression in florets, especially in pollen during anther development. Ugp1 silencing by RNA interference or cosuppression results in male sterility. Expressing a double-stranded RNA interference construct in Ugp1-RI plants resulted in complete suppression of both Ugp1 and Ugp2, together with various pleiotropic developmental abnormalities, suggesting that UGPase plays critical roles in plant growth and development. More importantly, Ugp1-cosuppressing plants contained unprocessed intron-containing primary transcripts derived from transcription of the overexpression construct. These aberrant transcripts undergo temperature-sensitive splicing in florets, leading to a novel thermosensitive genic male sterility. Pollen mother cells (PMCs) of Ugp1-silenced plants appeared normal before meiosis, but during meiosis, normal callose deposition was disrupted. Consequently, the PMCs began to degenerate at the early meiosis stage, eventually resulting in complete pollen collapse. In addition, the degeneration of the tapetum and middle layer was inhibited. These results demonstrate that rice Ugp1 is required for callose deposition during PMC meiosis and bridges the apoplastic unloading pathway and pollen development.
Figures

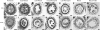


References
-
- Abe, T., Niiyama, H., and Sasahara, T. (2002). Cloning of cDNA for UDP-glucose pyrophosphorylase and the expression of mRNA in rice endosperm. Theor. Appl. Genet. 105 216–221. - PubMed
-
- Baulcombe, D. (2004). RNA silencing in plants. Nature 431 356–363. - PubMed
-
- Beclin, C., Boutet, S., Waterhouse, P., and Vaucheret, H. (2002). A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12 684–688. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

