Chaperoning ribonucleoprotein biogenesis in health and disease
- PMID: 17401408
- PMCID: PMC1852756
- DOI: 10.1038/sj.embor.7400941
Chaperoning ribonucleoprotein biogenesis in health and disease
Abstract
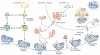
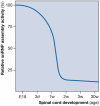
The survival motor neuron (SMN) protein is part of a macromolecular complex that functions in the biogenesis of small nuclear ribonucleoproteins (snRNPs)--the essential components of the pre-messenger RNA splicing machinery--as well as probably other RNPs. Reduced levels of SMN expression cause the inherited motor neuron disease spinal muscular atrophy (SMA). Knowledge of the composition, interactions and functions of the SMN complex has advanced greatly in recent years. The emerging picture is that the SMN complex acts as a macromolecular chaperone of RNPs to increase the efficiency and fidelity of RNA-protein interactions in vivo, and to provide an opportunity for these interactions to be regulated. In addition, it seems that RNA metabolism deficiencies underlie SMA. Here, a dual dysfunction hypothesis is presented in which two mechanistically and temporally distinct defects--that are dependent on the extent of SMN reduction in SMA--affect the homeostasis of specific messenger RNAs encoding proteins essential for motor neuron development and function.
Figures


References
-
- Battle DJ, Lau CK, Wan L, Deng H, Lotti F, Dreyfuss G (2006) The Gemin5 protein of the SMN complex identifies snRNAs. Mol Cell 23: 273–279 - PubMed
-
- Buhler D, Raker V, Luhrmann R, Fischer U (1999) Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum Mol Genet 8: 2351–2357 - PubMed
-
- Carissimi C, Baccon J, Straccia M, Chiarella P, Maiolica A, Sawyer A, Rappsilber J, Pellizzoni L (2005) Unrip is a component of SMN complexes active in snRNP assembly. FEBS Lett 579: 2348–2354 - PubMed
-
- Carissimi C, Saieva L, Baccon J, Chiarella P, Maiolica A, Sawyer A, Rappsilber J, Pellizzoni L (2006a) Gemin8 is a novel component of the survival motor neuron complex and functions in snRNP assembly. J Biol Chem 281: 8126–8134 - PubMed
-
- Carissimi C, Saieva L, Gabanella F, Pellizzoni L (2006b) Gemin8 is required for the architecture and function of the survival motor neuron complex. J Biol Chem 281: 37009–37016 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

