Expansion of adult beta-cell mass in response to increased metabolic demand is dependent on HNF-4alpha
- PMID: 17403778
- PMCID: PMC1838528
- DOI: 10.1101/gad.1535507
Expansion of adult beta-cell mass in response to increased metabolic demand is dependent on HNF-4alpha
Abstract
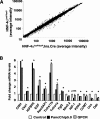
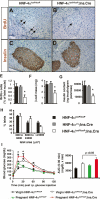
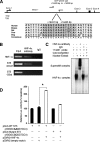
The failure to expand functional pancreatic beta-cell mass in response to increased metabolic demand is a hallmark of type 2 diabetes. Lineage tracing studies indicate that replication of existing beta-cells is the principle mechanism for beta-cell expansion in adult mice. Here we demonstrate that the proliferative response of beta-cells is dependent on the orphan nuclear receptor hepatocyte nuclear factor-4alpha (HNF-4alpha), the gene that is mutated in Maturity-Onset Diabetes of the Young 1 (MODY1). Computational analysis of microarray expression profiles from isolated islets of mice lacking HNF-4alpha in pancreatic beta-cells reveals that HNF-4alpha regulates selected genes in the beta-cell, many of which are involved in proliferation. Using a physiological model of beta-cell expansion, we show that HNF-4alpha is required for beta-cell replication and the activation of the Ras/ERK signaling cascade in islets. This phenotype correlates with the down-regulation of suppression of tumorigenicity 5 (ST5) in HNF-4alpha mutants, which we identify as a novel regulator of ERK phosphorylation in beta-cells and a direct transcriptional target of HNF-4alpha in vivo. Together, these results indicate that HNF-4alpha is essential for the physiological expansion of adult beta-cell mass in response to increased metabolic demand.
Figures


References
-
- Ahlgren U., Jonsson J., Jonsson L., Simu K., Edlund H., Jonsson J., Jonsson L., Simu K., Edlund H., Jonsson L., Simu K., Edlund H., Simu K., Edlund H., Edlund H. β-Cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the β-cell phenotype and maturity onset diabetes. Genes & Dev. 1998;12:1763–1768. - PMC - PubMed
-
- Amaral M.E., Ueno M., Carvalheira J.B., Carneiro E.M., Velloso L.A., Saad M.J., Boschero A.C., Ueno M., Carvalheira J.B., Carneiro E.M., Velloso L.A., Saad M.J., Boschero A.C., Carvalheira J.B., Carneiro E.M., Velloso L.A., Saad M.J., Boschero A.C., Carneiro E.M., Velloso L.A., Saad M.J., Boschero A.C., Velloso L.A., Saad M.J., Boschero A.C., Saad M.J., Boschero A.C., Boschero A.C. Prolactin-signal transduction in neonatal rat pancreatic islets and interaction with the insulin-signaling pathway. Horm. Metab. Res. 2003;35:282–289. - PubMed
-
- Amaral M.E., Cunha D.A., Anhe G.F., Ueno M., Carneiro E.M., Velloso L.A., Bordin S., Boschero A.C., Cunha D.A., Anhe G.F., Ueno M., Carneiro E.M., Velloso L.A., Bordin S., Boschero A.C., Anhe G.F., Ueno M., Carneiro E.M., Velloso L.A., Bordin S., Boschero A.C., Ueno M., Carneiro E.M., Velloso L.A., Bordin S., Boschero A.C., Carneiro E.M., Velloso L.A., Bordin S., Boschero A.C., Velloso L.A., Bordin S., Boschero A.C., Bordin S., Boschero A.C., Boschero A.C. Participation of prolactin receptors and phosphatidylinositol 3-kinase and MAP kinase pathways in the increase in pancreatic islet mass and sensitivity to glucose during pregnancy. J. Endocrinol. 2004;183:469–476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
