HLA-B-associated transcript 3 (Bat3)/Scythe is essential for p300-mediated acetylation of p53
- PMID: 17403783
- PMCID: PMC1838535
- DOI: 10.1101/gad.1534107
HLA-B-associated transcript 3 (Bat3)/Scythe is essential for p300-mediated acetylation of p53
Abstract
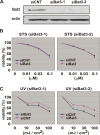
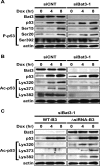
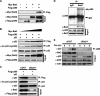
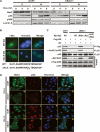
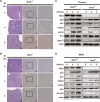
In response to DNA damage, p53 undergoes post-translational modifications (including acetylation) that are critical for its transcriptional activity. However, the mechanism by which p53 acetylation is regulated is still unclear. Here, we describe an essential role for HLA-B-associated transcript 3 (Bat3)/Scythe in controlling the acetylation of p53 required for DNA damage responses. Depletion of Bat3 from human and mouse cells markedly impairs p53-mediated transactivation of its target genes Puma and p21. Although DNA damage-induced phosphorylation, stabilization, and nuclear accumulation of p53 are not significantly affected by Bat3 depletion, p53 acetylation is almost completely abolished. Bat3 forms a complex with p300, and an increased amount of Bat3 enhances the recruitment of p53 to p300 and facilitates subsequent p53 acetylation. In contrast, Bat3-depleted cells show reduced p53-p300 complex formation and decreased p53 acetylation. Furthermore, consistent with our in vitro findings, thymocytes from Bat3-deficient mice exhibit reduced induction of puma and p21, and are resistant to DNA damage-induced apoptosis in vivo. Our data indicate that Bat3 is a novel and essential regulator of p53-mediated responses to genotoxic stress, and that Bat3 controls DNA damage-induced acetylation of p53.
Figures

References
-
- Ahn J., Prives C., Prives C. The C-terminus of p53: The more you learn the less you know. Nat. Struct. Biol. 2001;8:730–732. - PubMed
-
- Appella E., Anderson C.W., Anderson C.W. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 2001;268:2764–2772. - PubMed
-
- Avantaggiati M.L., Ogryzko V., Gardner K., Giordano A., Levine A.S., Kelly K., Ogryzko V., Gardner K., Giordano A., Levine A.S., Kelly K., Gardner K., Giordano A., Levine A.S., Kelly K., Giordano A., Levine A.S., Kelly K., Levine A.S., Kelly K., Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. - PubMed
-
- Ayed A., Mulder F.A., Yi G.S., Lu Y., Kay L.E., Arrowsmith C.H., Mulder F.A., Yi G.S., Lu Y., Kay L.E., Arrowsmith C.H., Yi G.S., Lu Y., Kay L.E., Arrowsmith C.H., Lu Y., Kay L.E., Arrowsmith C.H., Kay L.E., Arrowsmith C.H., Arrowsmith C.H. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 2001;8:756–760. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
