Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo
- PMID: 17404621
- PMCID: PMC1838939
- DOI: 10.1172/JCI30117
Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo
Abstract
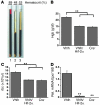
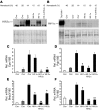
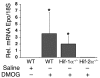
Erythropoiesis is critically dependent on erythropoietin (EPO), a glycoprotein hormone that is regulated by hypoxia-inducible factor (HIF). Hepatocytes are the primary source of extrarenal EPO in the adult and express HIF-1 and HIF-2, whose roles in the hypoxic induction of EPO remain controversial. In order to define the role of HIF-1 and HIF-2 in the regulation of hepatic EPO expression, we have generated mice with conditional inactivation of Hif-1alpha and/or Hif-2alpha (Epas1) in hepatocytes. We have previously shown that inactivation of the von Hippel-Lindau tumor suppressor pVHL, which targets both HIFs for proteasomal degradation, results in increased hepatic Epo production and polycythemia independent of Hif-1alpha. Here we show that conditional inactivation of Hif-2alpha in pVHL-deficient mice suppressed hepatic Epo and the development of polycythemia. Furthermore, we found that physiological Epo expression in infant livers required Hif-2alpha but not Hif-1alpha and that the hypoxic induction of liver Epo in anemic adults was Hif-2alpha dependent. Since other Hif target genes such phosphoglycerate kinase 1 (Pgk) were Hif-1alpha dependent, we provide genetic evidence that HIF-1 and HIF-2 have distinct roles in the regulation of hypoxia-inducible genes and that EPO is preferentially regulated by HIF-2 in the liver.
Figures
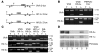

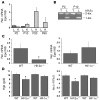
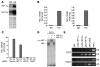
Comment in
-
HIF-1 and HIF-2: working alone or together in hypoxia?J Clin Invest. 2007 Apr;117(4):862-5. doi: 10.1172/JCI31750. J Clin Invest. 2007. PMID: 17404612 Free PMC article.
References
-
- Ebert B.L., Bunn H.F. Regulation of the erythropoietin gene. Blood. 1999;94:1864–1877. - PubMed
-
- Wu H., Lee S.H., Gao J., Liu X., Iruela-Arispe M.L. Inactivation of erythropoietin leads to defects in cardiac morphogenesis. Development. 1999;126:3597–3605. - PubMed
-
- Wu H., Liu X., Jaenisch R., Lodish H.F. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell. 1995;83:59–67. - PubMed
-
- Dame C., et al. Erythropoietin mRNA expression in human fetal and neonatal tissue. Blood. 1998;92:3218–3225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

