Structure and evolution of the Ivy protein family, unexpected lysozyme inhibitors in Gram-negative bacteria
- PMID: 17405861
- PMCID: PMC1847508
- DOI: 10.1073/pnas.0611019104
Structure and evolution of the Ivy protein family, unexpected lysozyme inhibitors in Gram-negative bacteria
Abstract
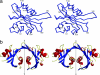
Part of an ancestral bactericidal system, vertebrate C-type lysozyme targets the peptidoglycan moiety of bacterial cell walls. We report the crystal structure of a protein inhibitor of C-type lysozyme, the Escherichia coli Ivy protein, alone and in complex with hen egg white lysozyme. Ivy exhibits a novel fold in which a protruding five-residue loop appears essential to its inhibitory effect. This feature guided the identification of Ivy orthologues in other Gram-negative bacteria. The structure of the evolutionary distant Pseudomonas aeruginosa Ivy orthologue was also determined in complex with hen egg white lysozyme, and its antilysozyme activity was confirmed. Ivy expression protects porous cell-wall E. coli mutants from the lytic effect of lysozyme, suggesting that it is a response against the permeabilizing effects of the innate vertebrate immune system. As such, Ivy acts as a virulence factor for a number of Gram-negative bacteria-infecting vertebrates.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


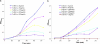
References
-
- MacFarlane G. Alexander Fleming: The Man and the Myth. Oxford, UK: Oxford Univ Press; 1985.
-
- Prager EM, Jollès P. Animal Lysozymes c and g: An Overview. Switzerland: Birkhäuser, Basel; 1996. - PubMed
-
- Blake CC, Koenig DF, Mair GA, North AC, Phillips DC, Sarma VR. Nature. 1965;206:757–761. - PubMed
-
- Holtje JV. Exs. 1996;75:105–110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

