Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions
- PMID: 17406506
- PMCID: PMC2752288
- DOI: 10.1038/nprot.2006.204
Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions
Abstract
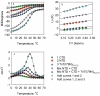
Circular dichroism (CD) is an excellent spectroscopic technique for following the unfolding and folding of proteins as a function of temperature. One of its principal applications is to determine the effects of mutations and ligands on protein and polypeptide stability. If the change in CD as a function of temperature is reversible, analysis of the data may be used to determined the van't Hoff enthalpy and entropy of unfolding, the midpoint of the unfolding transition and the free energy of unfolding. Binding constants of protein-protein and protein-ligand interactions may also be estimated from the unfolding curves. Analysis of CD spectra obtained as a function of temperature is also useful to determine whether a protein has unfolding intermediates. Measurement of the spectra of five folded proteins and their unfolding curves at a single wavelength requires approximately 8 h.
Figures

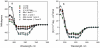

References
-
- Velluz L, Legrand M, Grosjean M. Optical Circular DIchroism: Principles, Masurements, and Applications. Verlag Chemie Academic Press Inc; New York and London: 1965.
-
- Beychok S. Circular dichroism of biological macromolecules. Science. 1966;154:1288–99. - PubMed
-
- Adler AJ, Greenfield NJ, Fasman GD. Circular dichroism and optical rotatory dispersion of proteins and polypeptides. Methods Enzymol. 1973;27:675–735. - PubMed
-
- Johnson WC., Jr. Protein secondary structure and circular dichroism: a practical guide. Proteins. 1990;7:205–14. - PubMed
-
- Woody RW. Circular dichroism. Methods Enzymol. 1995;246:34–71. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

