Using circular dichroism spectra to estimate protein secondary structure
- PMID: 17406547
- PMCID: PMC2728378
- DOI: 10.1038/nprot.2006.202
Using circular dichroism spectra to estimate protein secondary structure
Abstract
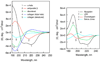
Circular dichroism (CD) is an excellent tool for rapid determination of the secondary structure and folding properties of proteins that have been obtained using recombinant techniques or purified from tissues. The most widely used applications of protein CD are to determine whether an expressed, purified protein is folded, or if a mutation affects its conformation or stability. In addition, it can be used to study protein interactions. This protocol details the basic steps of obtaining and interpreting CD data, and methods for analyzing spectra to estimate the secondary structural composition of proteins. CD has the advantage that measurements may be made on multiple samples containing < or =20 microg of proteins in physiological buffers in a few hours. However, it does not give the residue-specific information that can be obtained by x-ray crystallography or NMR.
Figures

References
-
- Velluz L, Legrand M, Grosjean M. Optical Circular DIchroism: Principles, Masurements, and Applications. New York and London: Verlag Chemie Academic Press Inc; 1965.
-
- Beychok S. Circular dichroism of biological macromolecules. Science. 1966;154:1288–1299. - PubMed
-
- Sreerama N, Woody RW. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 2004;383:318–351. - PubMed
-
- Holzwarth G, Doty P. The Ultraviolet Circular Dichroism of Polypeptides. J. Am. Chem. Soc. 1965;87:218–228. - PubMed
-
- Greenfield N, Fasman GD. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969;8:4108–4116. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

