Enzymatic cleavage specificity of the proalpha1(V) chain processing analysed by site-directed mutagenesis
- PMID: 17407447
- PMCID: PMC1904530
- DOI: 10.1042/BJ20070051
Enzymatic cleavage specificity of the proalpha1(V) chain processing analysed by site-directed mutagenesis
Abstract
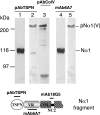
The proteolytic processing of procollagen V is complex and depends on the activity of several enzymes among which the BMP-1 (bone morphogenetic protein-1)/tolloid metalloproteinase and the furin-like proprotein convertases. Few of these processing interactions could have been predicted by analysing the presence of conserved consensus sequences in the proalpha1(V) chain. In the present study we opted for a cell approach that allows a straightforward identification of processing interactions. A construct encompassing the complete N-terminal end of the proalpha1(V) chain, referred to as Nalpha1, was recombinantly expressed to be used for enzymatic assays and for antibody production. Structural analysis showed that Nalpha1 is a monomer composed of a compact globule and an extended tail, which correspond respectively to the non-collagenous Nalpha1 subdomains, TSPN-1 (thrombospondin-1 N-terminal domain-like) and the variable region. Nalpha1 was efficiently cleaved by BMP-1 indicating that the triple helix is not required for enzyme activity. By mutating residues flanking the cleavage site, we showed that the aspartate residue at position P2' is essential for BMP-1 activity. BMP-1 activity at the C-terminal end of the procollagen V was assessed by generating a furin double mutant (R1584A/R1585A). We showed that, in absence of furin activity, BMP-1 is capable of processing the C-propeptide even though less efficiently than furin. Altogether, our results provide new relevant information on this complex and poorly understood mechanism of enzymatic processing in procollagen V function.
Figures






References
-
- Ricard-Blum S., Ruggiero F. The collagen superfamily: from the extracellular matrix to the cell membrane. Pathol. Biol. 2005;53:30–42. - PubMed
-
- Prockop D. J., Sieron A. L., Li S. W. Procollagen N-proteinase and procollagen C-proteinase. Two unusual metalloproteinases that are essential for procollagen processing probably have important roles in development and cell signalling. Matrix Biol. 1998;16:399–408. - PubMed
-
- Sasaki T., Gohring W., Mann K., Brakebusch C., Yamada Y., Fassler R., Timpl R. Short arm region of laminin-5 gamma2 chain: structure, mechanism of processing and binding to heparin and proteins. J. Mol. Biol. 2001;314:751–763. - PubMed
-
- Colombo M., Brittingham R. J., Klement J. F., Majsterek I., Birk D. E., Uitto J., Fertala A. Procollagen VII self-assembly depends on site-specific interactions and is promoted by cleavage of the NC2 domain with procollagen C-proteinase. Biochemistry. 2003;42:11434–11442. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

