Improved residue contact prediction using support vector machines and a large feature set
- PMID: 17407573
- PMCID: PMC1852326
- DOI: 10.1186/1471-2105-8-113
Improved residue contact prediction using support vector machines and a large feature set
Abstract
Background: Predicting protein residue-residue contacts is an important 2D prediction task. It is useful for ab initio structure prediction and understanding protein folding. In spite of steady progress over the past decade, contact prediction remains still largely unsolved.
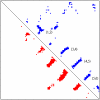
Results: Here we develop a new contact map predictor (SVMcon) that uses support vector machines to predict medium- and long-range contacts. SVMcon integrates profiles, secondary structure, relative solvent accessibility, contact potentials, and other useful features. On the same test data set, SVMcon's accuracy is 4% higher than the latest version of the CMAPpro contact map predictor. SVMcon recently participated in the seventh edition of the Critical Assessment of Techniques for Protein Structure Prediction (CASP7) experiment and was evaluated along with seven other contact map predictors. SVMcon was ranked as one of the top predictors, yielding the second best coverage and accuracy for contacts with sequence separation > or = 12 on 13 de novo domains.
Conclusion: We describe SVMcon, a new contact map predictor that uses SVMs and a large set of informative features. SVMcon yields good performance on medium- to long-range contact predictions and can be modularly incorporated into a structure prediction pipeline.
Figures
References
-
- Rost B, Liu J, Przybylski D, Nair R, Wrzeszczynski K, Bigelow H, Ofran Y. Prediction of protein structure through evolution. In: Gasteiger J, Engel T, editor. Handbook of Chemoinformatics – From Data to Knowledge. New York: Wiley; 2003. pp. 1789–1811.
-
- Olmea O, Rost B, Valencia A. Effective use of sequence correlation and conservation in fold recognition. J Mol Biol. 1999;295:1221–1239. - PubMed
-
- Cheng J, Baldi P. A Machine Learning Information Retrieval Approach to Protein Fold Recognition. Bioinformatics. 2006;22:1456–1463. - PubMed
-
- Aszodi A, Gradwell M, Taylor W. Global fold determination from a small number of distance restraints. J Mol Biol. 1995;251:308–326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

