From genes to functional classes in the study of biological systems
- PMID: 17407596
- PMCID: PMC1853114
- DOI: 10.1186/1471-2105-8-114
From genes to functional classes in the study of biological systems
Abstract
Background: With the popularization of high-throughput techniques, the need for procedures that help in the biological interpretation of results has increased enormously. Recently, new procedures inspired in systems biology criteria have started to be developed.
Results: Here we present FatiScan, a web-based program which implements a threshold-independent test for the functional interpretation of large-scale experiments that does not depend on the pre-selection of genes based on the multiple application of independent tests to each gene. The test implemented aims to directly test the behaviour of blocks of functionally related genes, instead of focusing on single genes. In addition, the test does not depend on the type of the data used for obtaining significance values, and consequently different types of biologically informative terms (gene ontology, pathways, functional motifs, transcription factor binding sites or regulatory sites from CisRed) can be applied to different classes of genome-scale studies. We exemplify its application in microarray gene expression, evolution and interactomics.
Conclusion: Methods for gene set enrichment which, in addition, are independent from the original data and experimental design constitute a promising alternative for the functional profiling of genome-scale experiments. A web server that performs the test described and other similar ones can be found at: http://www.babelomics.org.
Figures
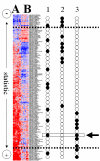
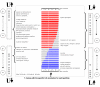
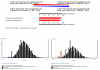
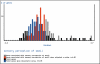
References
-
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. - PubMed
-
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. - PubMed
-
- Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. - PubMed
-
- Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

